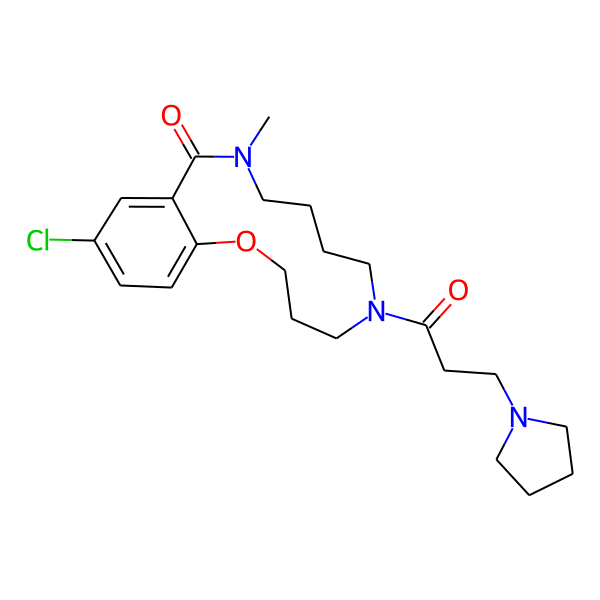
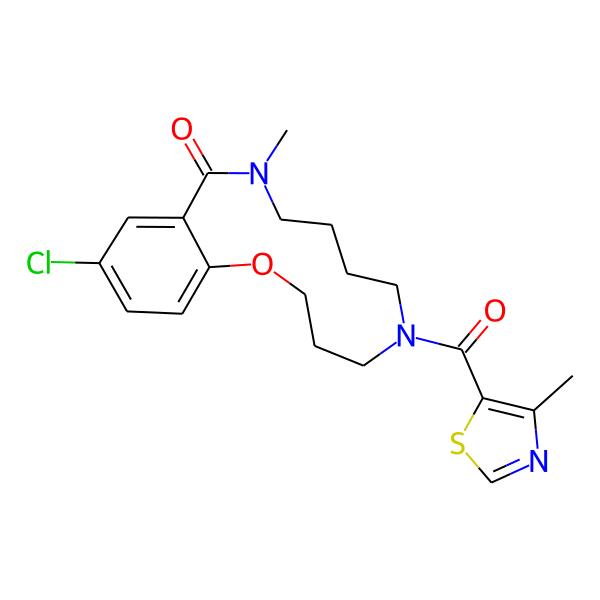
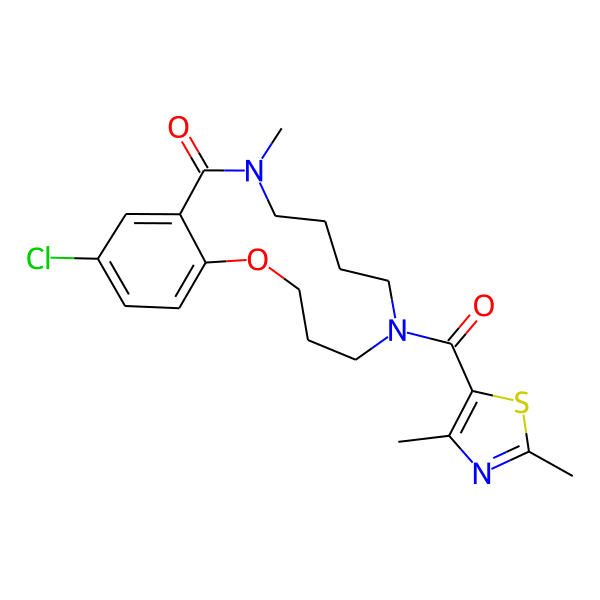
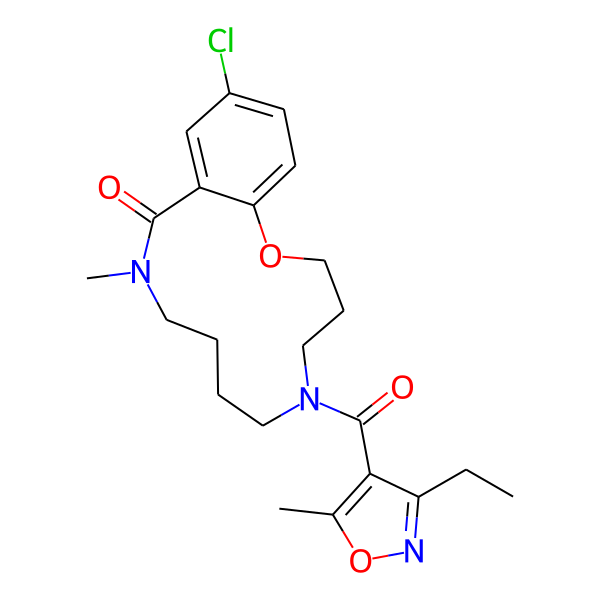
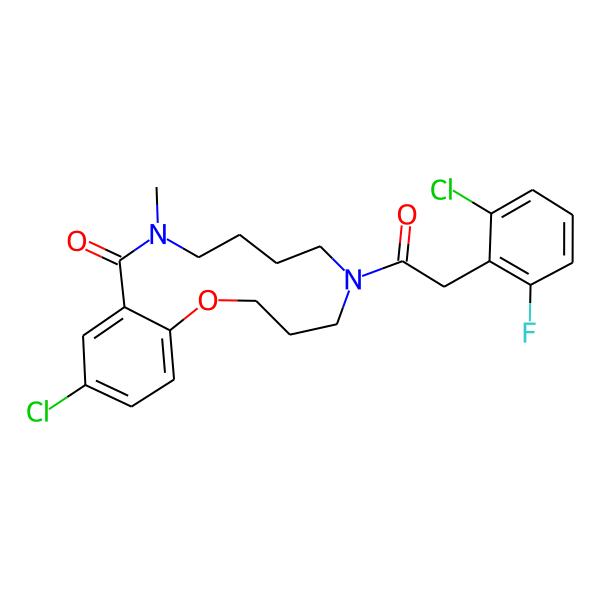
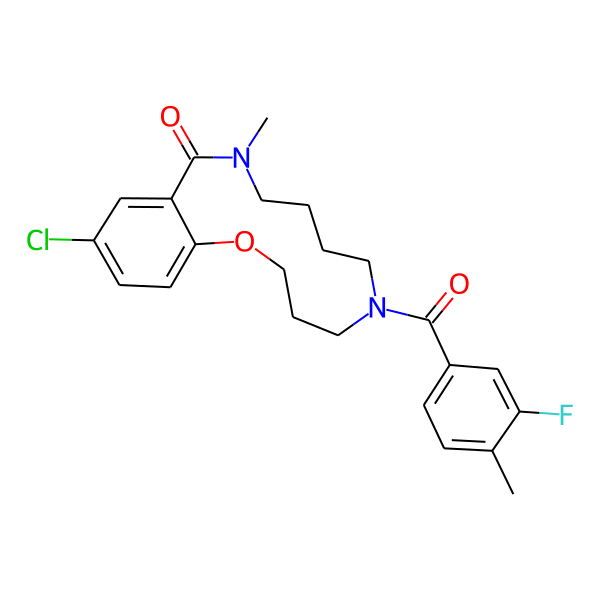
MC-1186
| Name | |||
|---|---|---|---|
| Unique ID | MC-1186 | ||
| Original ID | BAS_51616104 (Rzepiela et al., 2022) | ||
| Common Name | |||
| Structure Representations | |||
| InchiKey | NUHJXEVEWYDQNF-UHFFFAOYSA-N | ||
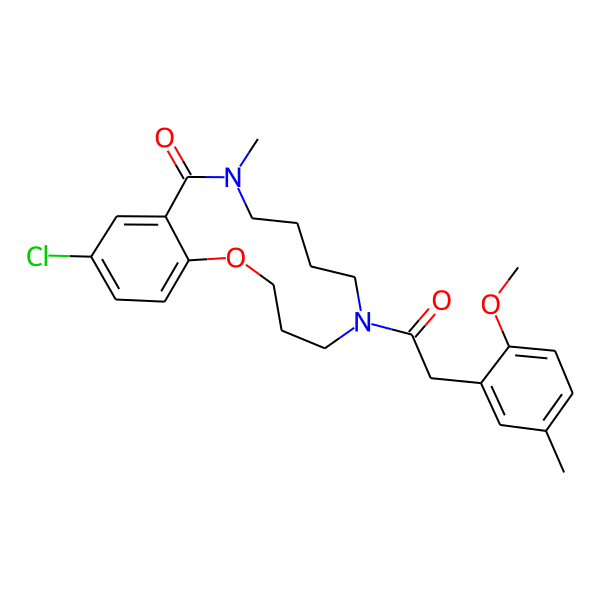
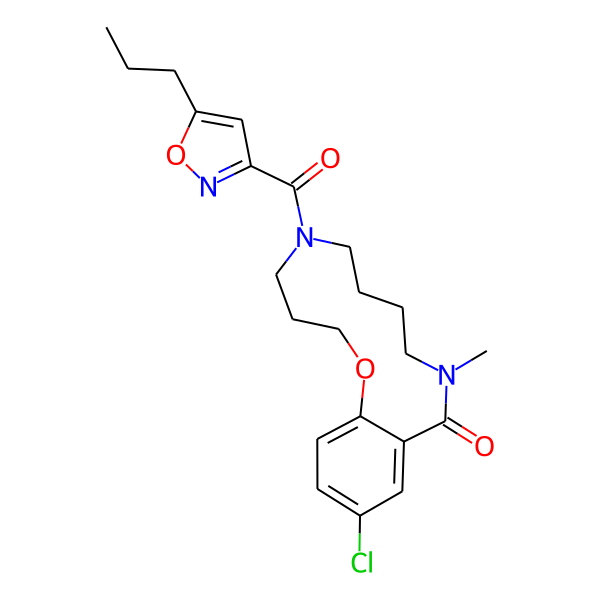
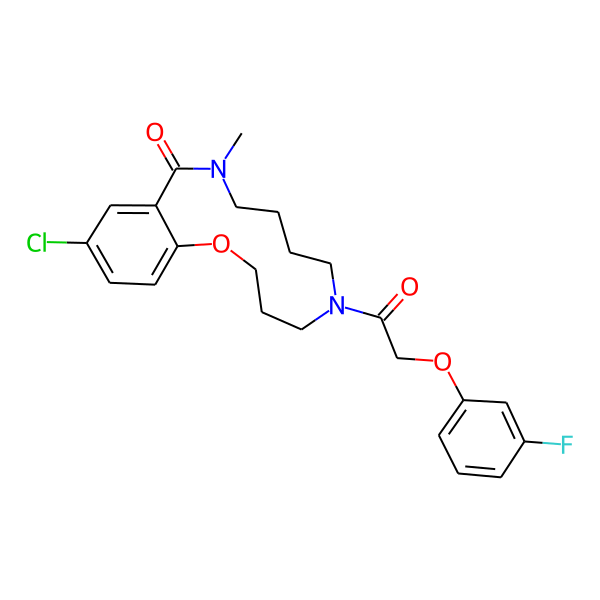
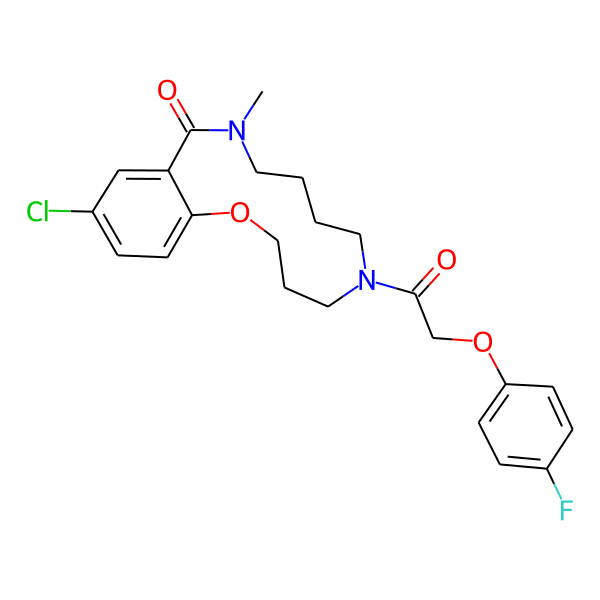
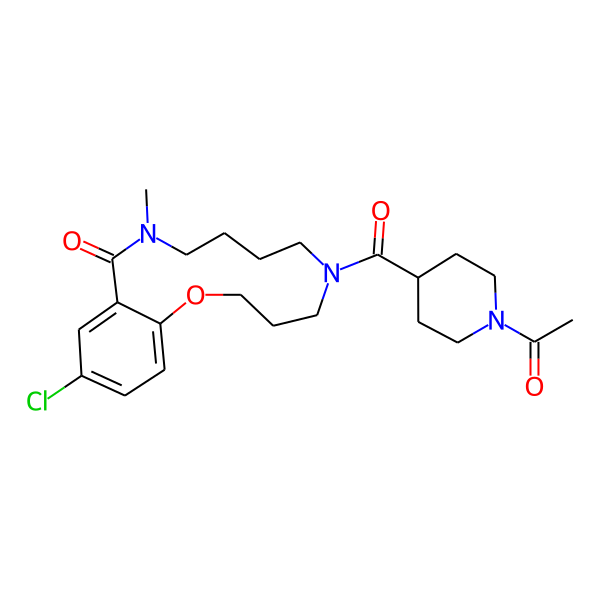
| Isomeric SMILES | COc1ccc(C)cc1CC(=O)N1CCCCN(C)C(=O)c2cc(Cl)ccc2OCCC1 | ||
| SMILES (Ring) | C1=COCCCNCCCCNC1 | ||
| Permeability | |||
| Assay | PAMPA | ||
| Endpoint | Log Peff | ||
| Value | -5.061 | ||
| Unit | |||
| Standardized Value | -5.06 | ||
| Molecule Descriptors | |||
| MW (Da) | 458.99 | NRotB | 3 |
| HBA | 4 | Kier Index (Φ) | 8.39 |
| HBD | 0 | AR | 0.23 |
| cLogP | 4.36 | Fsp3 | 0.44 |
| TPSA (Å2) | 59.08 | MRS | 13 |
| Unique ID: ID for each unique molecule in this dataset; Original ID: The name or ID in original source; Standaridized value: Logarithmic values for permeability values (unit 10-6 cm/s), original data with sign '<' or '>' were removed during data standardization | |||
| Abbreviations: MW (Da): Molecular Weight; HBA: Hydrogen bond acceptor; HBD: Hydrogen bond donor; cLogP: Calculated lipophilicity; TPSA: Topological polar surface area; NRotB: Number of rotatable bonds; Φ: Kier flexibility Index: Fsp3: fraction of sp3 carbon atoms; MRS: macrocyclic ring size; AR: Amide ratio; | |||
Back to Browse