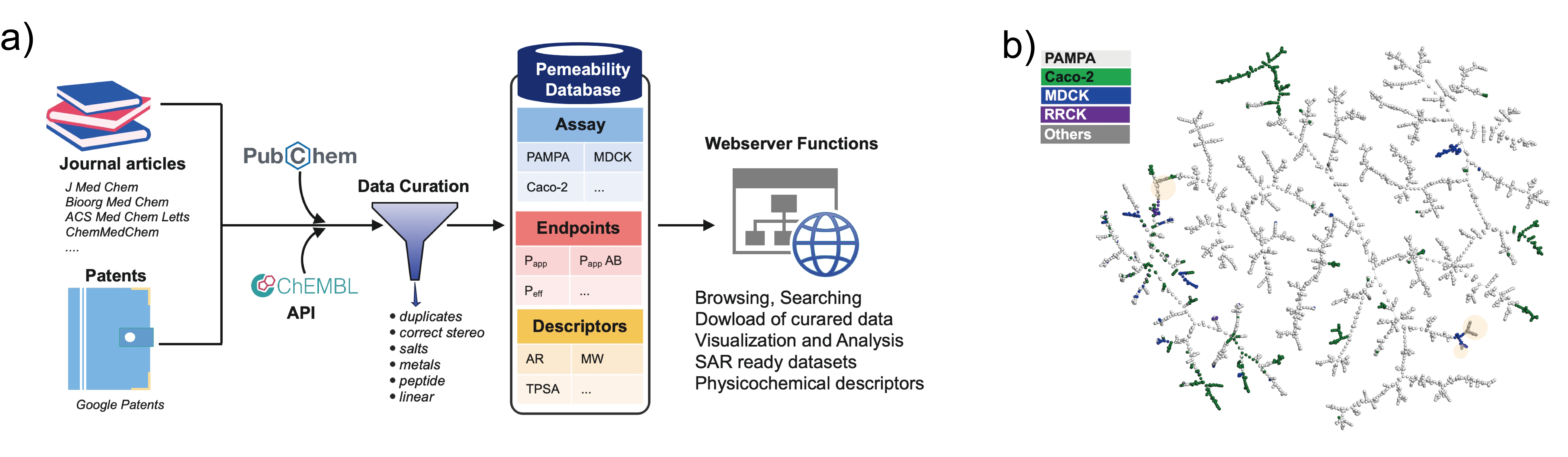
About macrocycles
Macrocycles, defined as rings of at least 12 heavy atoms, have garnered significant attention across various scientific fields, including drug discovery 1. Their appeal lies in their capacity to combine functional diversity and stereochemical complexity with conformational restriction to disc- and spherelike shapes. This distinctive structural attribute empowers macrocycles to bind with high affinity and selectivity to ‘difficult to drug’ targets that are challenging to modulate with traditional small-molecule drugs that adhere to the Rule of Five (Ro5) 1–3. Despite their size, macrocycles may still possess sufficient cell permeability and bioavailability, rendering them promising candidates for oral administration1,4. While macrocyclic drugs were historically derived from natural sources, there is a growing inclination towards de novo designed macrocycles among FDA approved drugs1. The rational design of potent, cell-permeable, orally available macrocyclic pharmaceuticals poses numerous challenges, particularly concerning synthesis and conformational prediction 5–7.
The process of passive cell permeability involves several steps, including desolvation when the drug transitions from the extracellular aqueous environment to interacting with the negatively charged phospholipid head groups before penetrating into the hydrophobic membrane interior (Figure 1). These steps are then reversed as the drug moves into the cytosol. Each of these processes is influenced to varying degrees by the molecular properties of the drug. For example, the compound's polarity, represented by its 3D polar surface area (PSA), significantly impacts the desolvation kinetics8. The compound's size, approximated by the radius of gyration (Rgyr), affects the rate of diffusion across the membrane, while its lipophilicity (cLogP or cLogD) is crucial for the thermodynamics of permeation8.