MC-0082
| Name | |||
|---|---|---|---|
| Unique ID | MC-0082 | ||
| Original ID | 62 (Pennington et al., 2021) | ||
| Common Name | |||
| Structure Representations | |||
| InchiKey | DIIBEFRRSSRSPB-UHFFFAOYSA-N | ||
| Isomeric SMILES | CC1COc2ccccc2C2CCC(CC2)OCC2C(NS(C)(=O)=O)CCCN12 | ||
| SMILES (Ring) | C1CCCOCCNCCOCC1 | ||
| Permeability | |||
| Assay | MDCK | ||
| Endpoint | ER | ||
| Value | 22 | ||
| Unit | |||
| Standardized Value | 22.00 | ||
| Molecule Descriptors | |||
| MW (Da) | 422.59 | NRotB | 2 |
| HBA | 5 | Kier Index (Φ) | 6.49 |
| HBD | 1 | AR | 0.00 |
| cLogP | 2.89 | Fsp3 | 0.73 |
| TPSA (Å2) | 76.25 | MRS | 13 |
| Unique ID: ID for each unique molecule in this dataset; Original ID: The name or ID in original source; Standaridized value: Logarithmic values for permeability values (unit 10-6 cm/s), original data with sign '<' or '>' were removed during data standardization | |||
| Abbreviations: MW (Da): Molecular Weight; HBA: Hydrogen bond acceptor; HBD: Hydrogen bond donor; cLogP: Calculated lipophilicity; TPSA: Topological polar surface area; NRotB: Number of rotatable bonds; Φ: Kier flexibility Index: Fsp3: fraction of sp3 carbon atoms; MRS: macrocyclic ring size; AR: Amide ratio; | |||
| Select | Name | Structure | MW(Da) | HBA | HBD | cLogP | TPSA(Å2) | Kier Index (Φ) |
|---|---|---|---|---|---|---|---|---|
| MC-0046 | 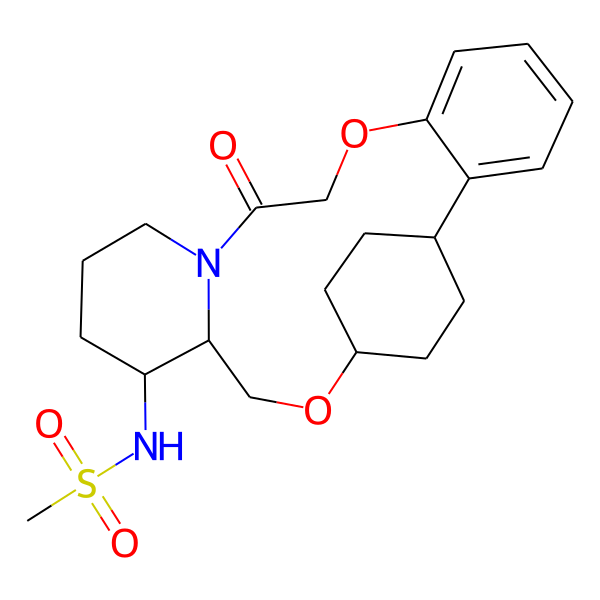 |
422.55 | 5 | 1 | 2.03 | 93.32 | 6.13 | |
| MC-0049 | 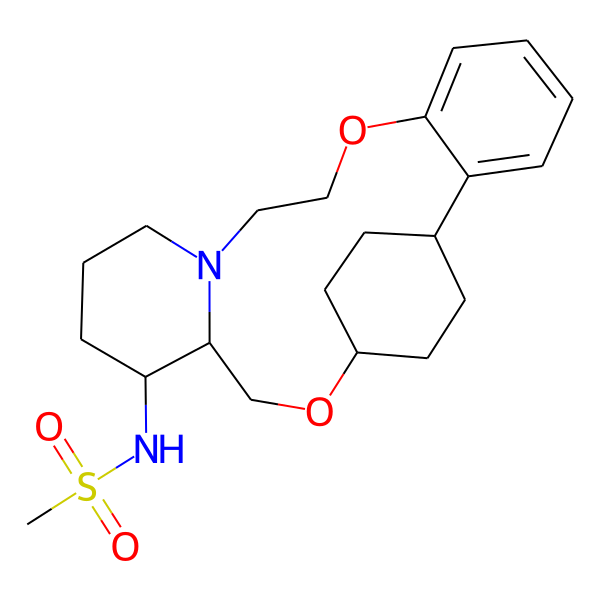 |
408.56 | 5 | 1 | 2.50 | 76.25 | 6.25 | |
| MC-0053 | 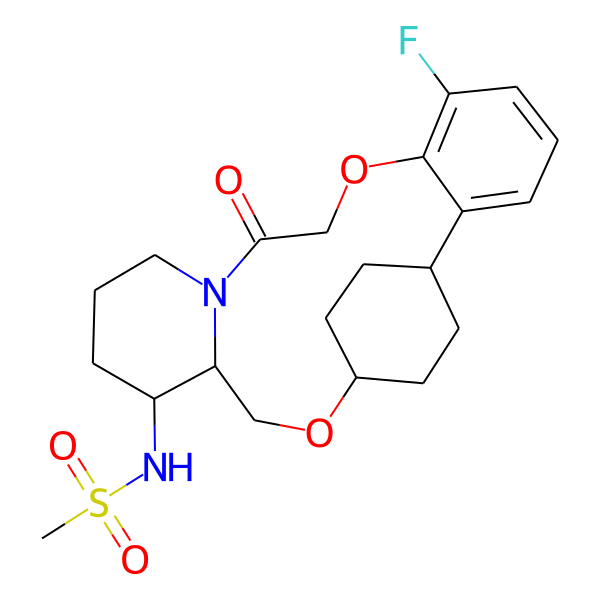 |
440.54 | 5 | 1 | 2.17 | 93.32 | 6.31 | |
| MC-0058 | 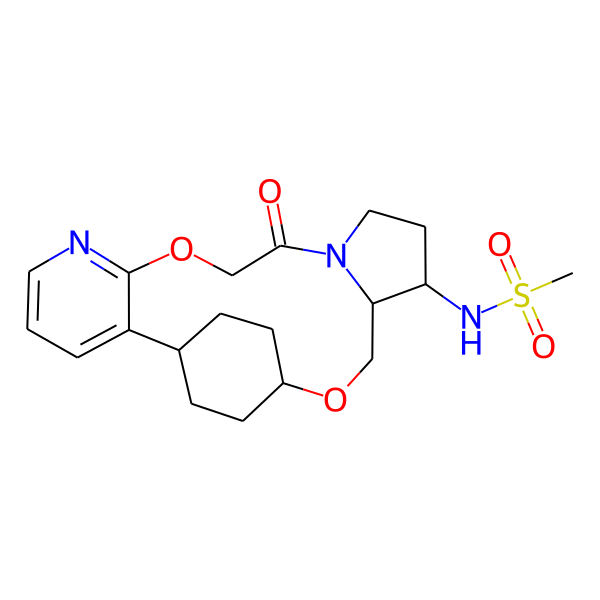 |
409.51 | 6 | 1 | 1.04 | 106.21 | 5.57 | |
| MC-0062 | 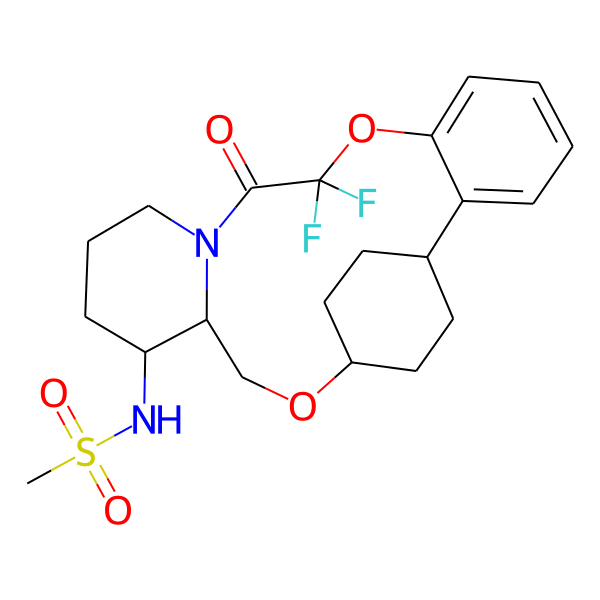 |
458.53 | 5 | 1 | 2.62 | 93.32 | 6.24 | |
| MC-0064 | 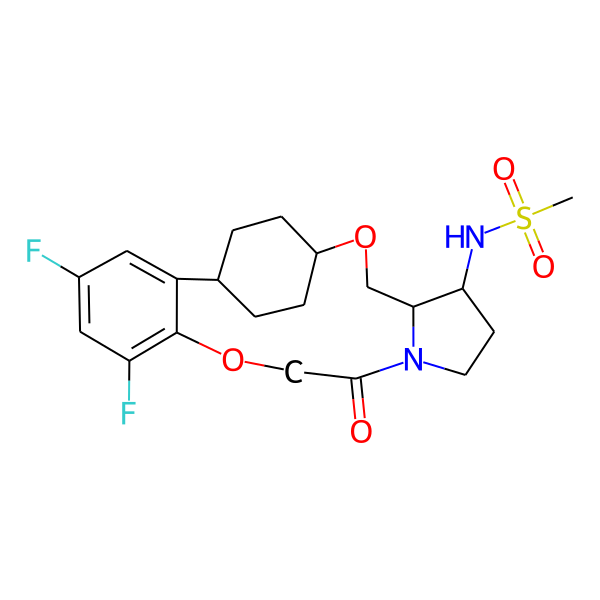 |
444.50 | 5 | 1 | 1.92 | 93.32 | 6.00 | |
| MC-0065 | 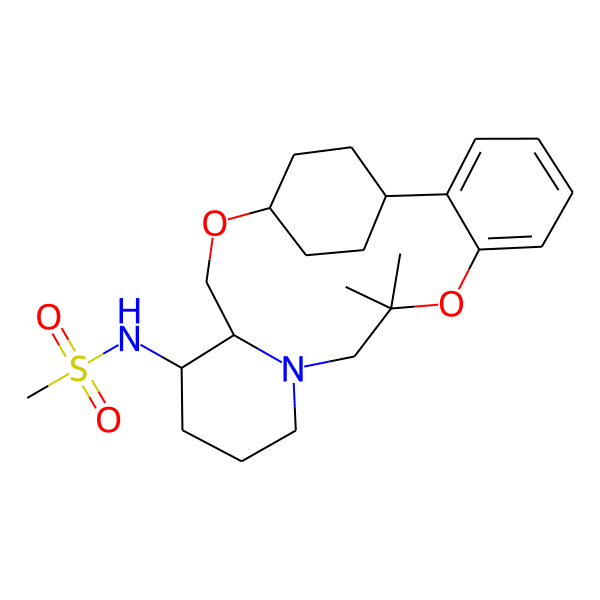 |
436.62 | 5 | 1 | 3.28 | 76.25 | 6.45 | |
| MC-0067 | 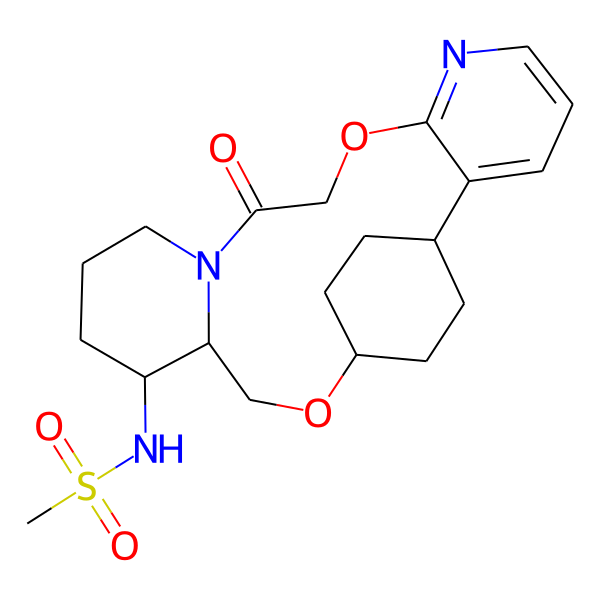 |
423.54 | 6 | 1 | 1.43 | 106.21 | 6.08 | |
| MC-0069 | 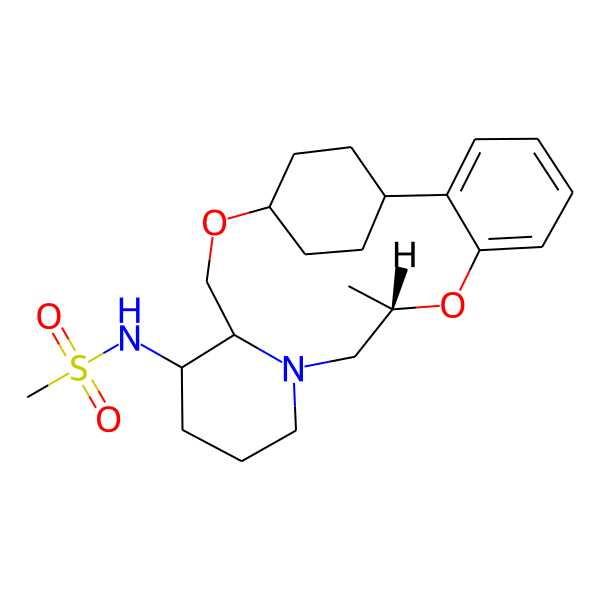 |
422.59 | 5 | 1 | 2.89 | 76.25 | 6.49 | |
| MC-0071 | 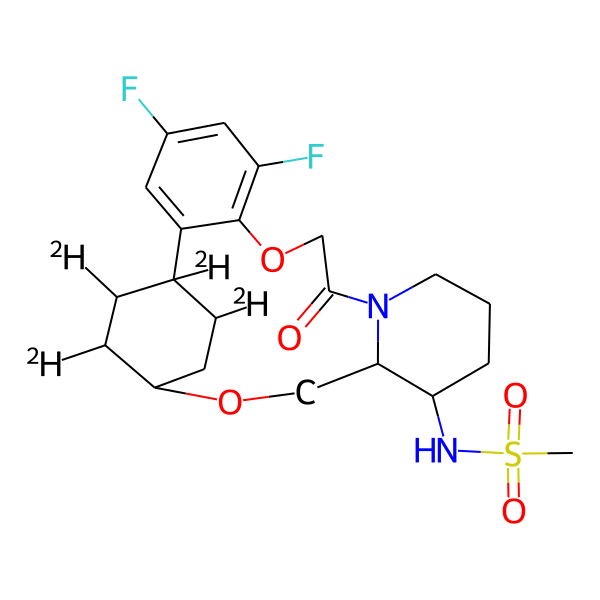 |
462.55 | 5 | 1 | 2.31 | 93.32 | 5.15 | |
| MC-0072 | 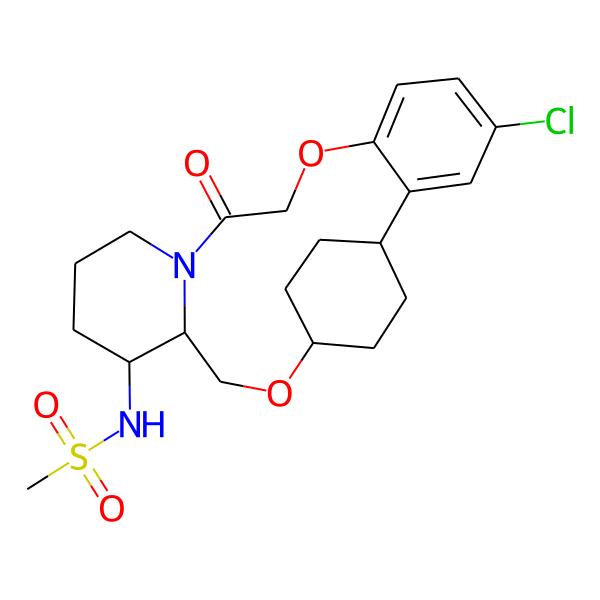 |
456.99 | 5 | 1 | 2.68 | 93.32 | 6.58 | |
| MC-0073 | 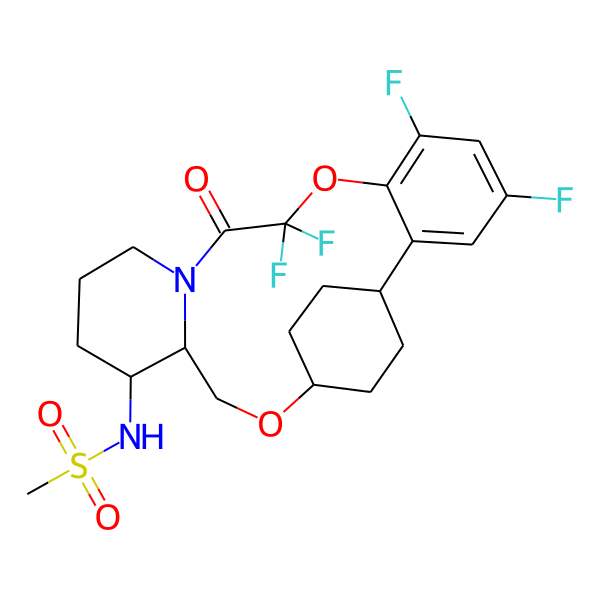 |
494.51 | 5 | 1 | 2.90 | 93.32 | 6.62 | |
| MC-0081 | 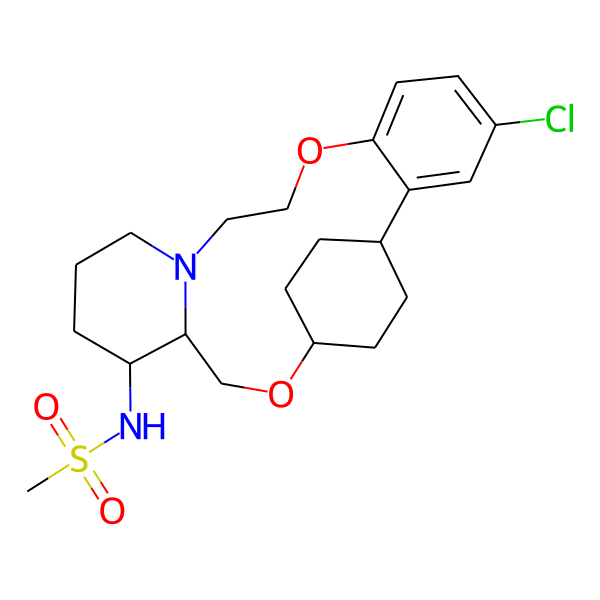 |
443.01 | 5 | 1 | 3.16 | 76.25 | 6.71 | |
| MC-0082 | 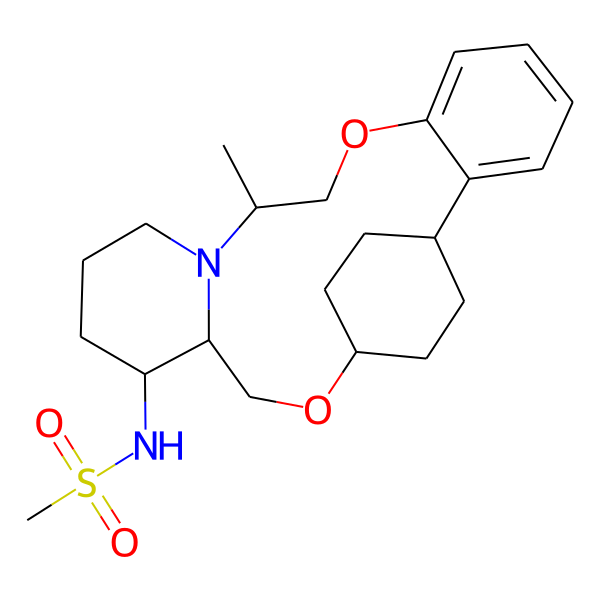 |
422.59 | 5 | 1 | 2.89 | 76.25 | 6.49 | |
| MC-0083 | 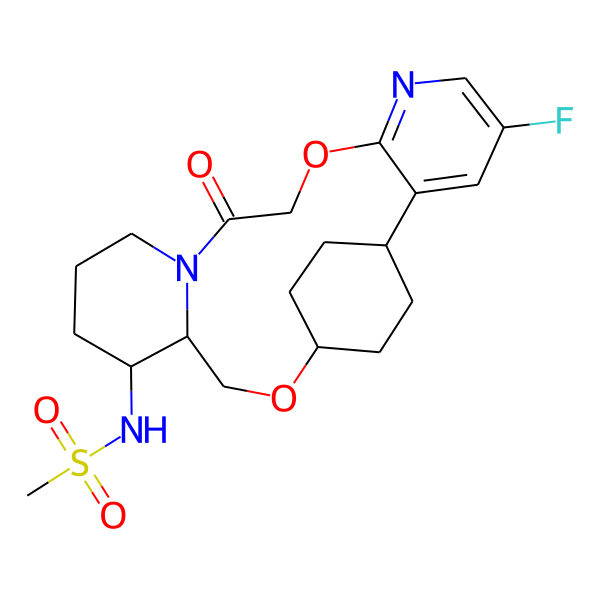 |
441.52 | 6 | 1 | 1.56 | 106.21 | 6.26 | |
| MC-4757 | 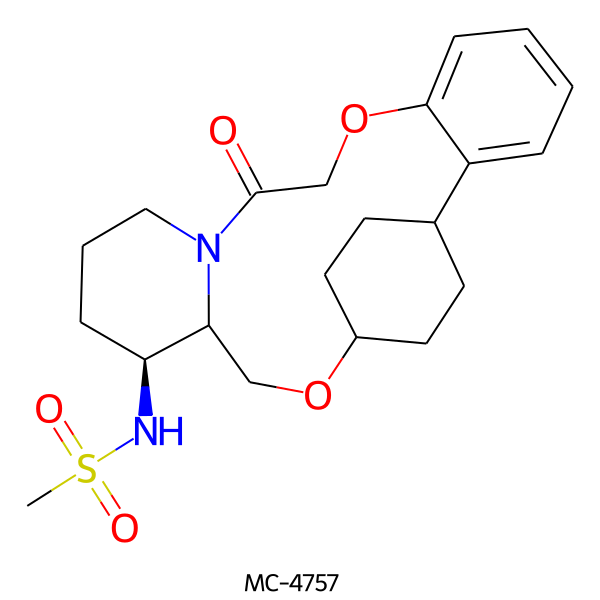 |
422.55 | 5 | 1 | 2.03 | 93.32 | 6.13 | |
| MC-4760 | 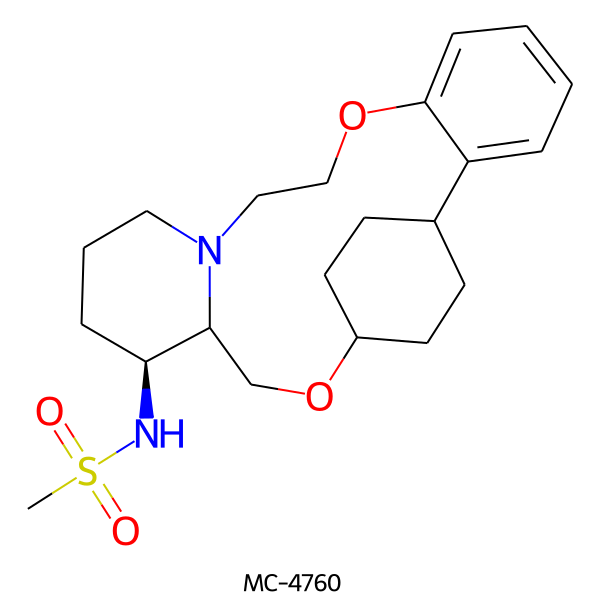 |
408.56 | 5 | 1 | 2.50 | 76.25 | 6.25 | |
| MC-4761 | 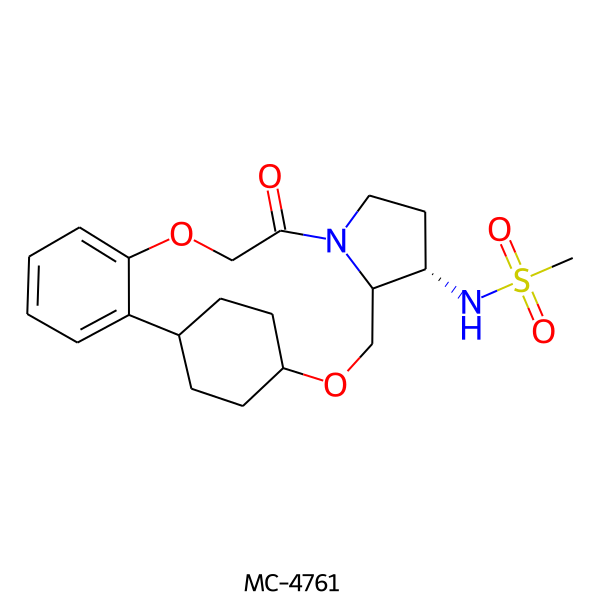 |
408.52 | 5 | 1 | 1.64 | 93.32 | 5.62 | |
| MC-4762 | 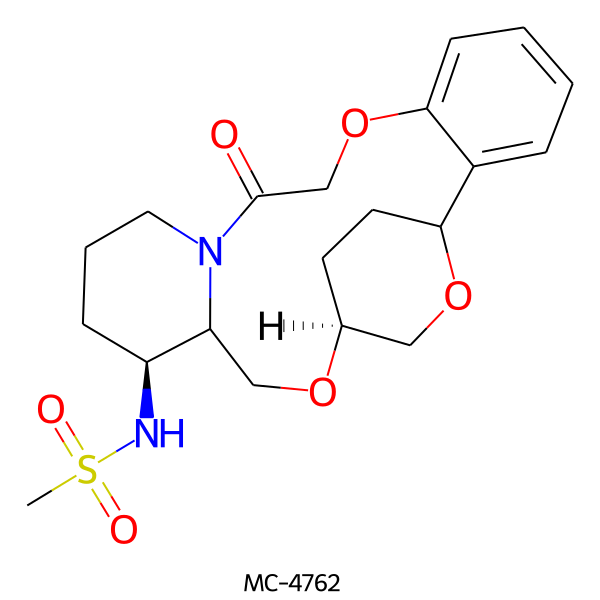 |
424.52 | 6 | 1 | 1.22 | 102.55 | 6.10 | |
| MC-4763 | 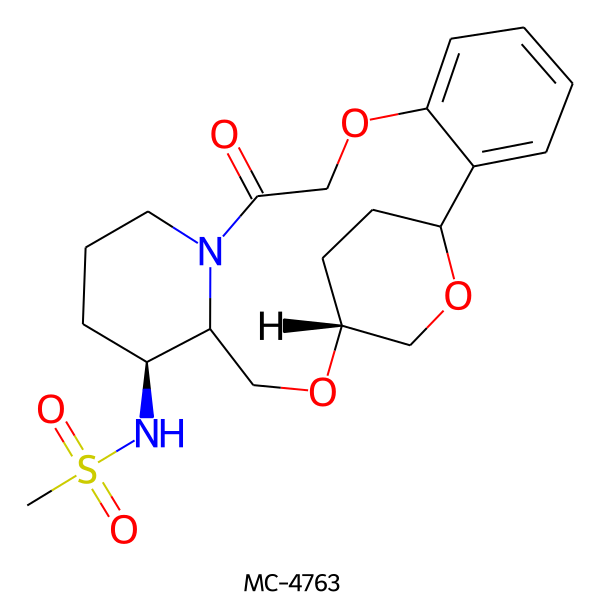 |
424.52 | 6 | 1 | 1.22 | 102.55 | 6.10 | |
| MC-4765 | 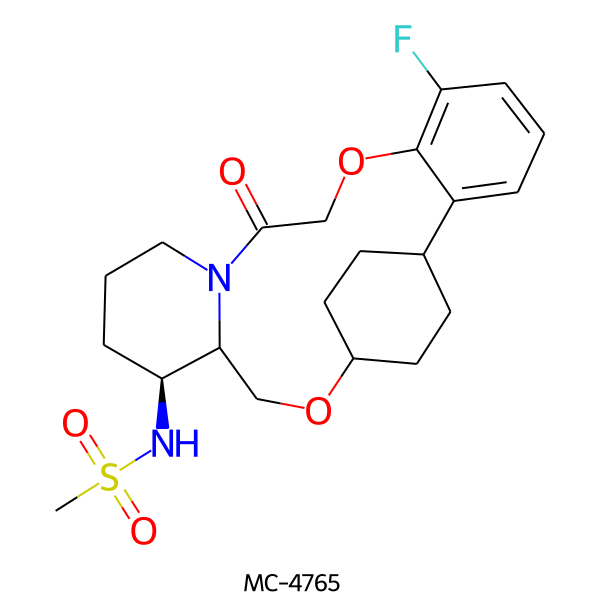 |
440.54 | 5 | 1 | 2.17 | 93.32 | 6.31 | |
| MC-4766 | 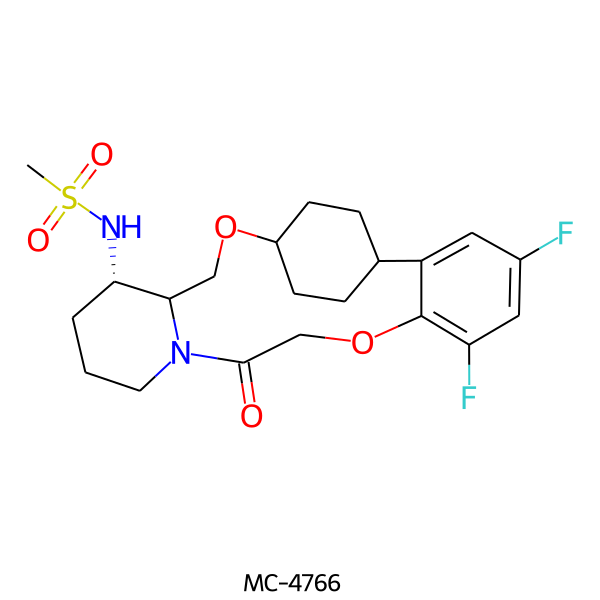 |
458.53 | 5 | 1 | 2.31 | 93.32 | 6.50 | |
| MC-4767 | 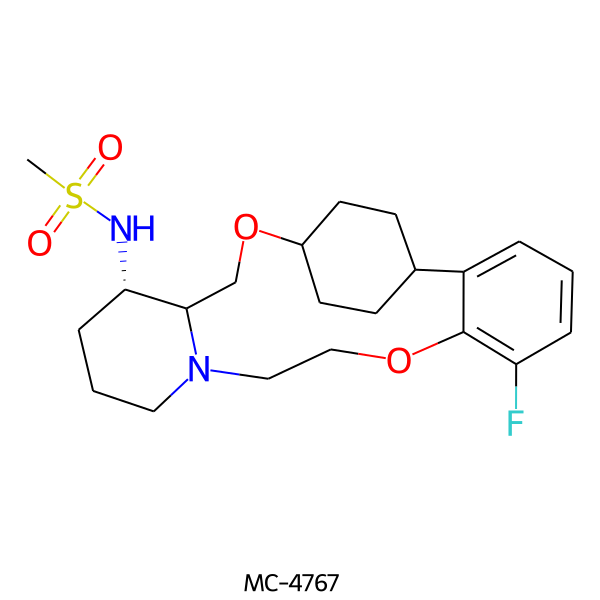 |
426.55 | 5 | 1 | 2.64 | 76.25 | 6.44 | |
| MC-4769 | 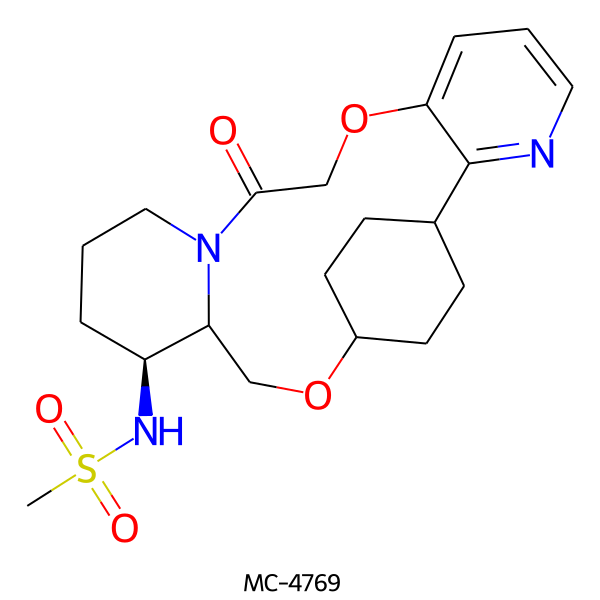 |
423.54 | 6 | 1 | 1.43 | 106.21 | 6.08 | |
| MC-4770 | 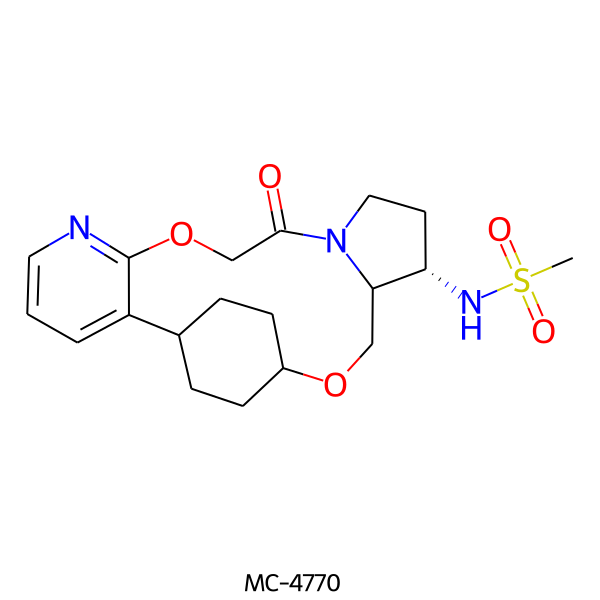 |
409.51 | 6 | 1 | 1.04 | 106.21 | 5.57 | |
| MC-4771 | 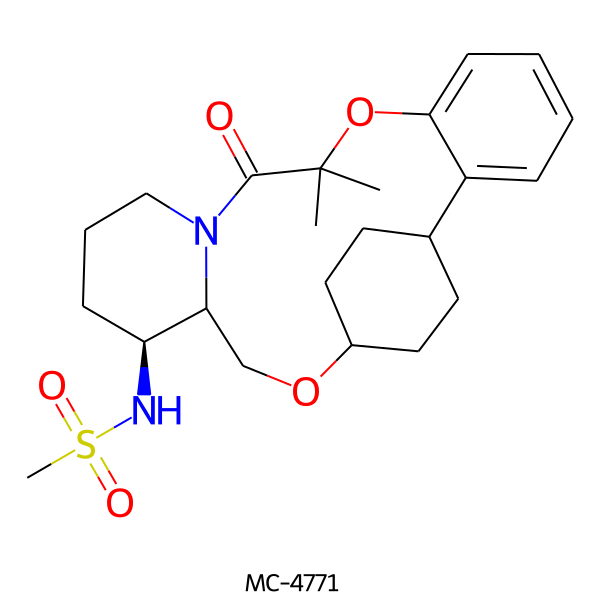 |
450.60 | 5 | 1 | 2.81 | 93.32 | 6.34 | |
| MC-4772 | 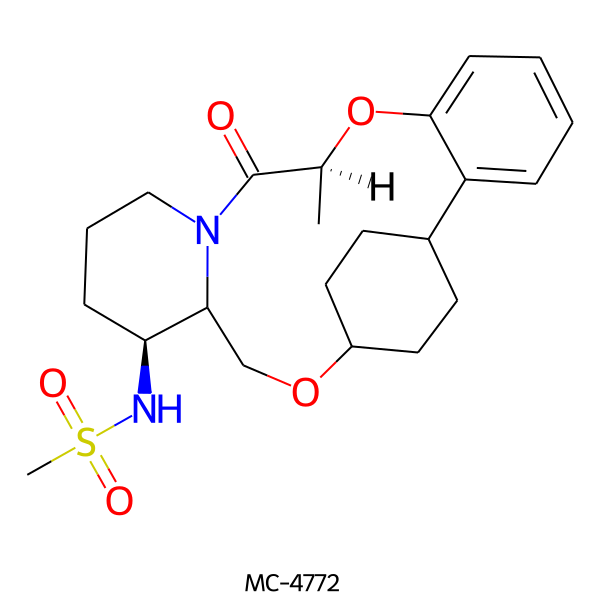 |
436.57 | 5 | 1 | 2.42 | 93.32 | 6.36 | |
| MC-4773 | 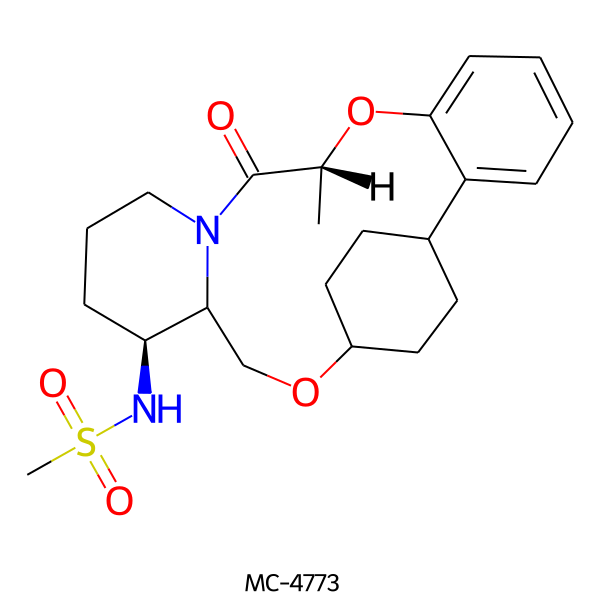 |
436.57 | 5 | 1 | 2.42 | 93.32 | 6.36 | |
| MC-4774 | 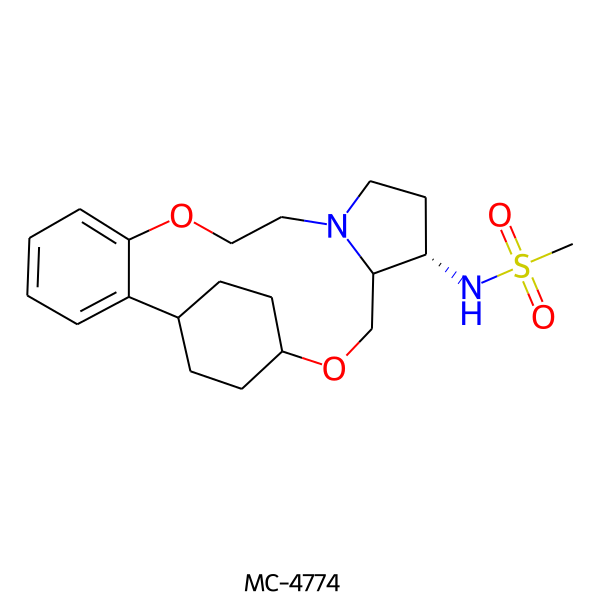 |
394.54 | 5 | 1 | 2.11 | 76.25 | 5.73 | |
| MC-4775 | 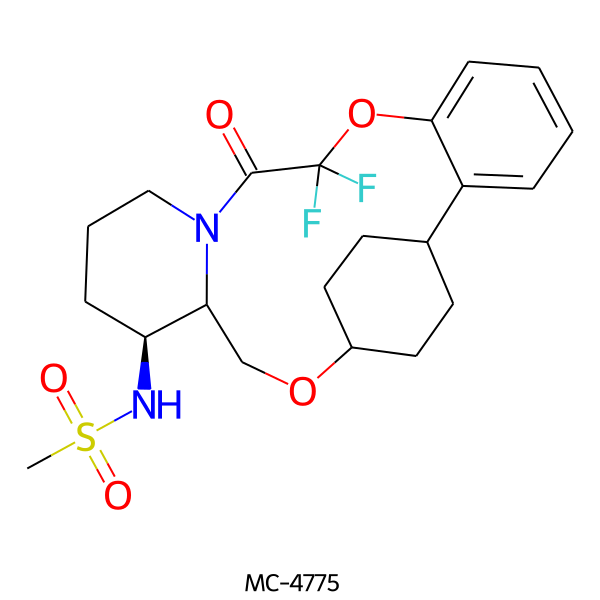 |
458.53 | 5 | 1 | 2.62 | 93.32 | 6.24 | |
| MC-4776 | 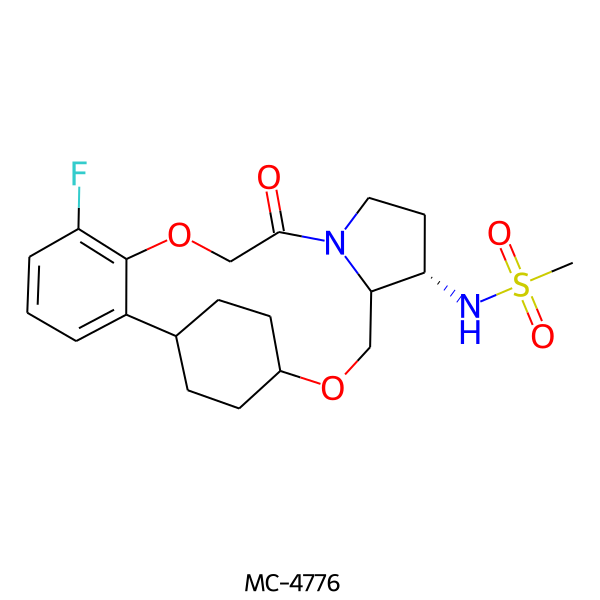 |
426.51 | 5 | 1 | 1.78 | 93.32 | 5.81 | |
| MC-4777 | 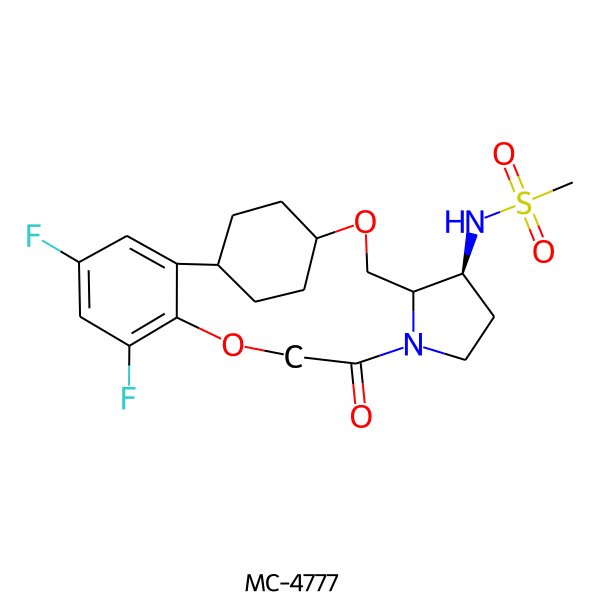 |
444.50 | 5 | 1 | 1.92 | 93.32 | 6.00 | |
| MC-4778 | 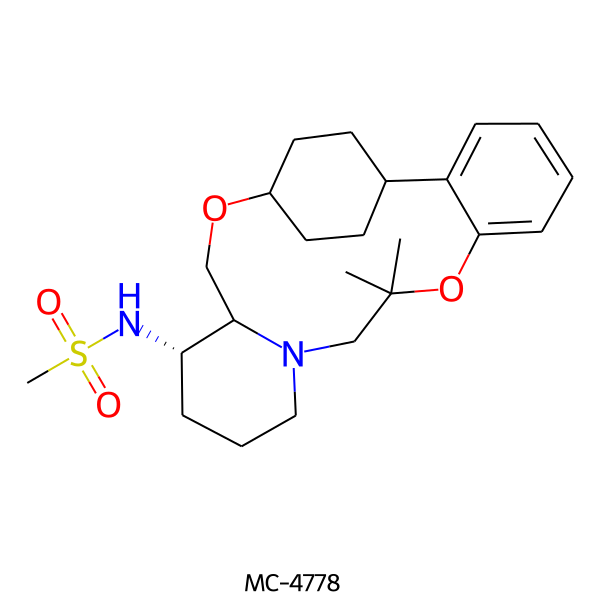 |
436.62 | 5 | 1 | 3.28 | 76.25 | 6.45 | |
| MC-4779 | 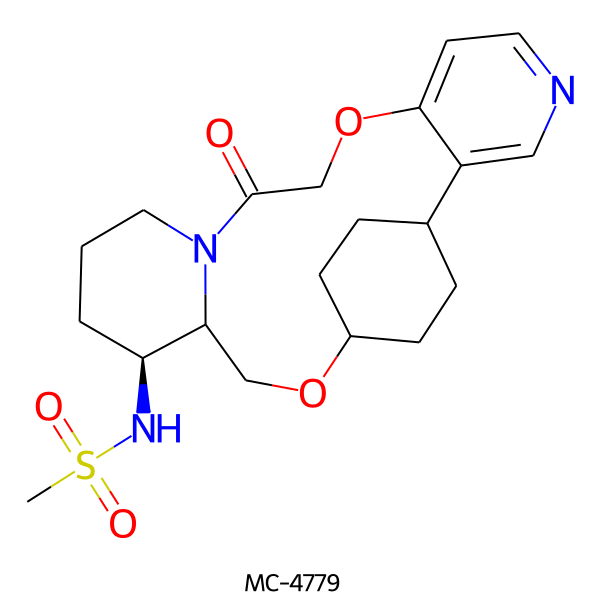 |
423.54 | 6 | 1 | 1.43 | 106.21 | 6.08 | |
| MC-4780 | 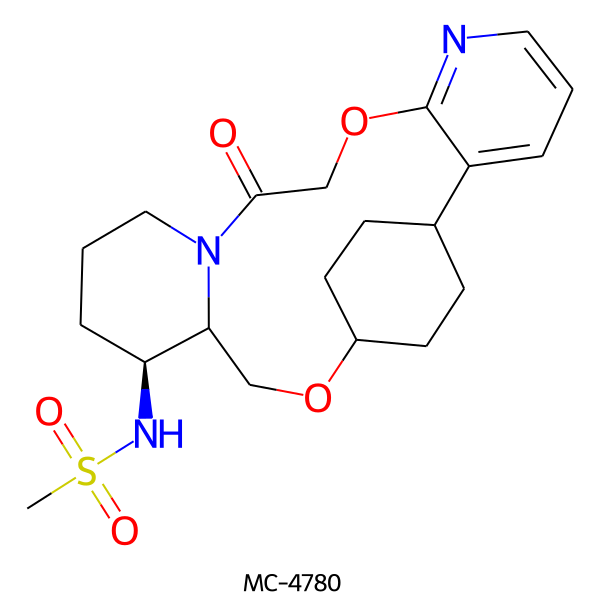 |
423.54 | 6 | 1 | 1.43 | 106.21 | 6.08 | |
| MC-4781 | 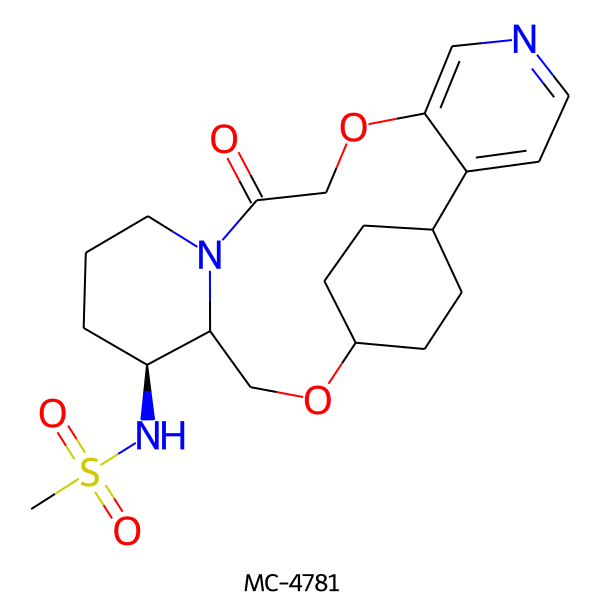 |
423.54 | 6 | 1 | 1.43 | 106.21 | 6.08 | |
| MC-4782 | 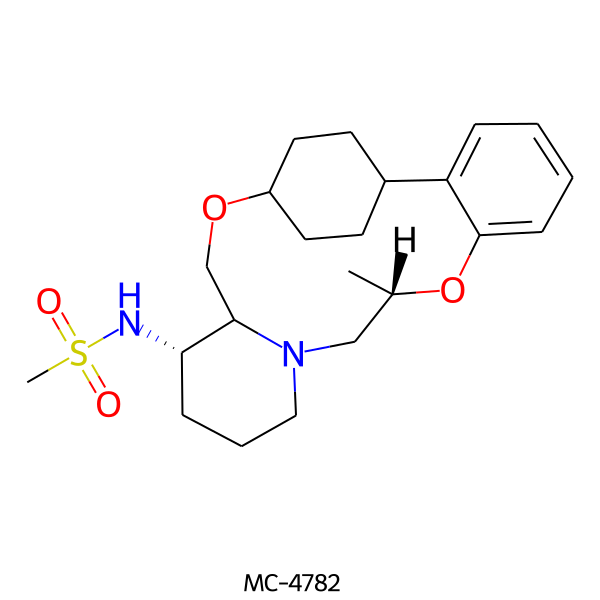 |
422.59 | 5 | 1 | 2.89 | 76.25 | 6.49 | |
| MC-4783 | 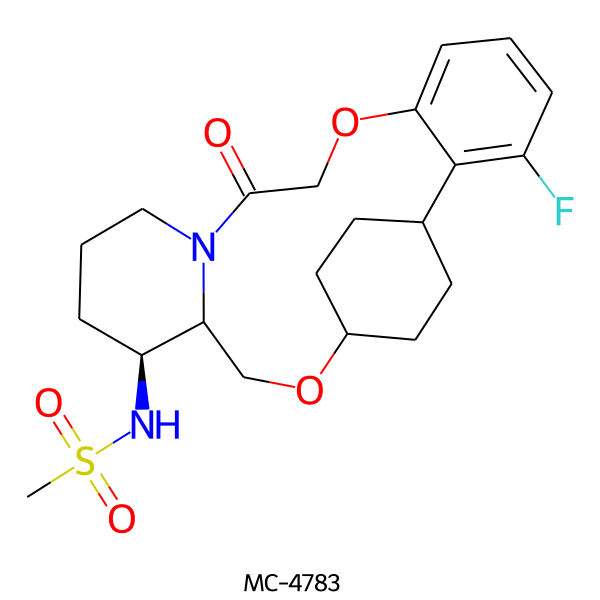 |
440.54 | 5 | 1 | 2.17 | 93.32 | 6.31 | |
| MC-4784 | 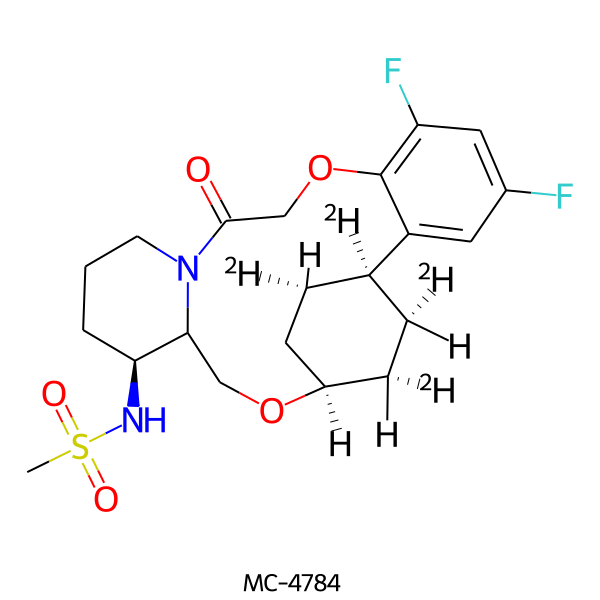 |
462.55 | 5 | 1 | 2.31 | 93.32 | 5.15 | |
| MC-4785 | 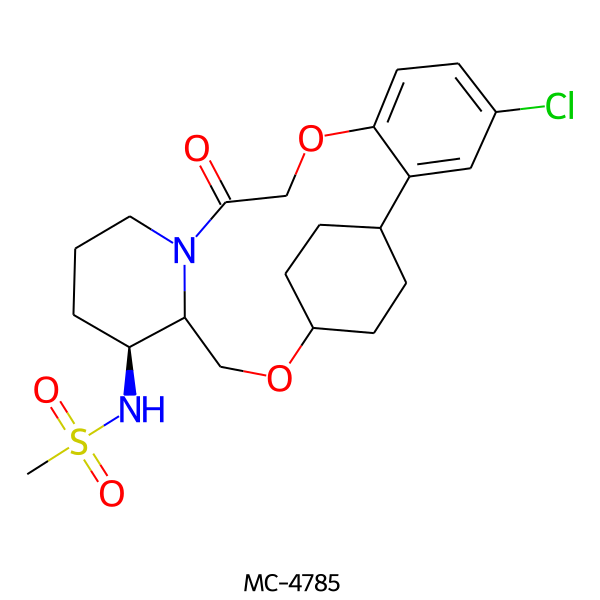 |
456.99 | 5 | 1 | 2.68 | 93.32 | 6.58 | |
| MC-4786 | 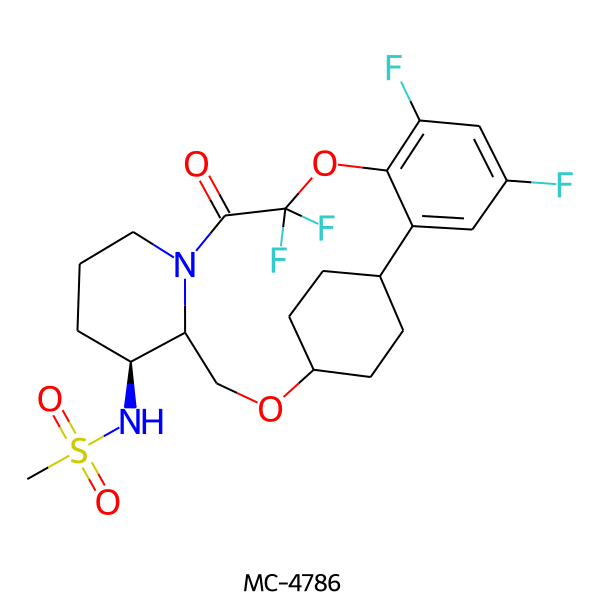 |
494.51 | 5 | 1 | 2.90 | 93.32 | 6.62 | |
| MC-4787 | 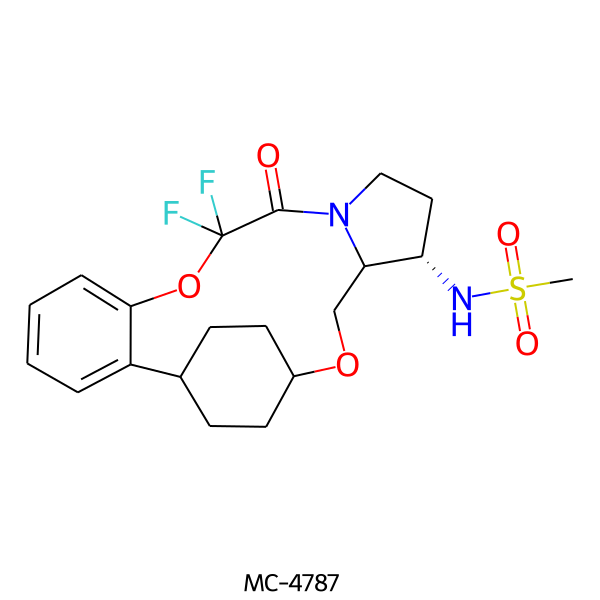 |
444.50 | 5 | 1 | 2.23 | 93.32 | 5.75 | |
| MC-4788 | 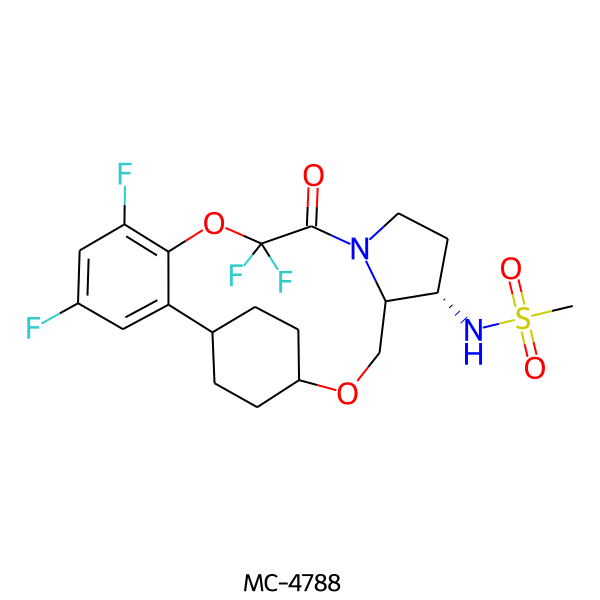 |
480.48 | 5 | 1 | 2.51 | 93.32 | 6.14 | |
| MC-4789 | 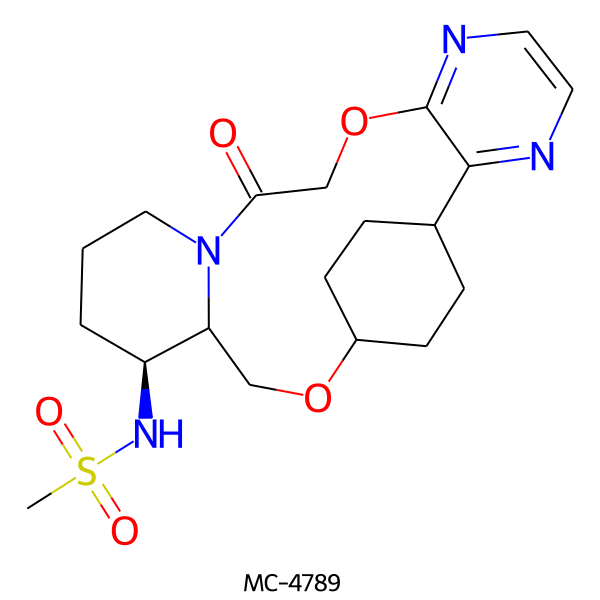 |
424.52 | 7 | 1 | 0.82 | 119.10 | 6.03 | |
| MC-4790 | 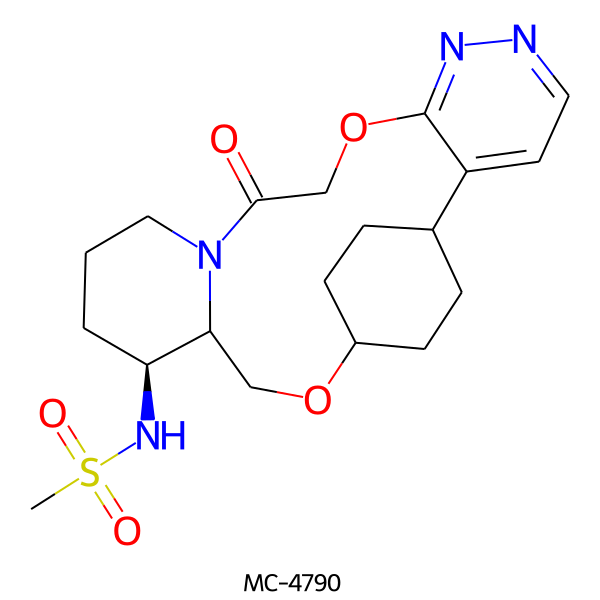 |
424.52 | 7 | 1 | 0.82 | 119.10 | 6.03 | |
| MC-4791 | 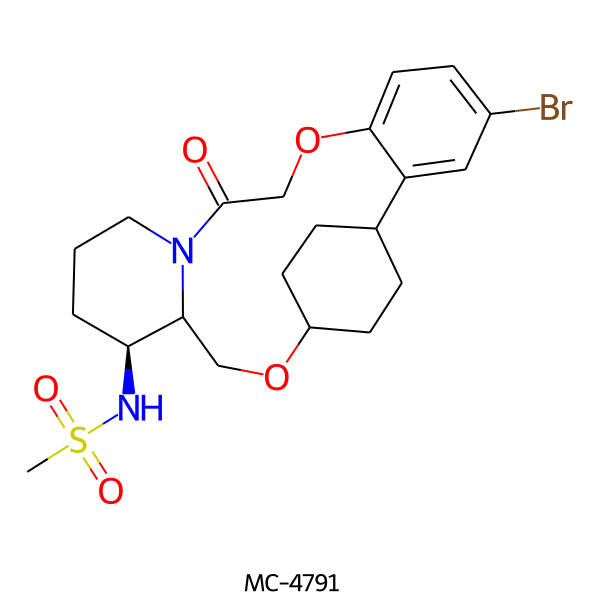 |
501.44 | 5 | 1 | 2.79 | 93.32 | 6.72 | |
| MC-4792 | 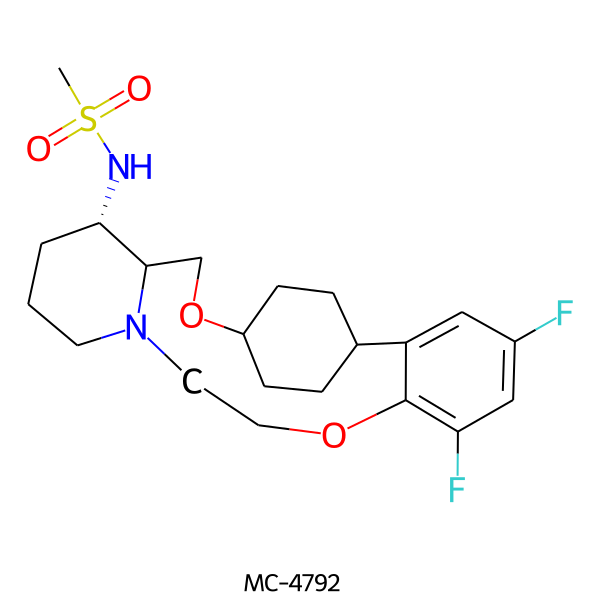 |
444.54 | 5 | 1 | 2.78 | 76.25 | 6.62 | |
| MC-4793 | 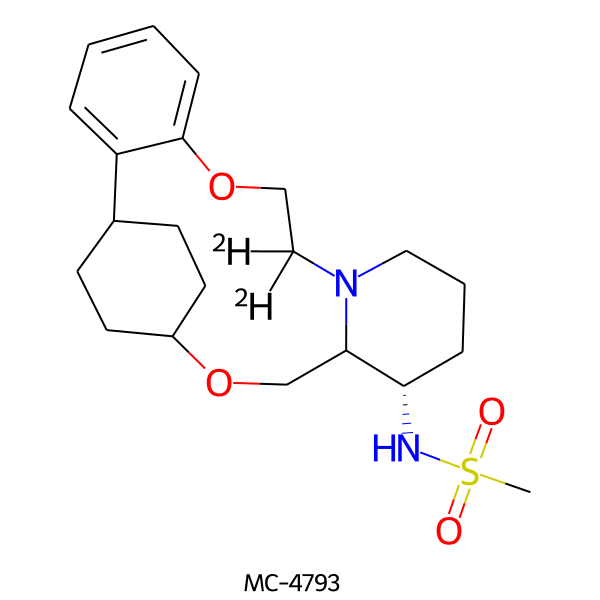 |
410.58 | 5 | 1 | 2.50 | 76.25 | 5.49 | |
| MC-4794 | 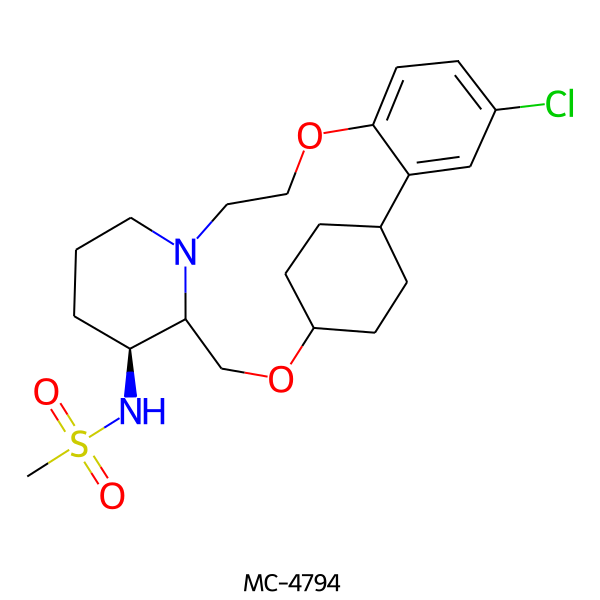 |
443.01 | 5 | 1 | 3.16 | 76.25 | 6.71 | |
| MC-4795 | 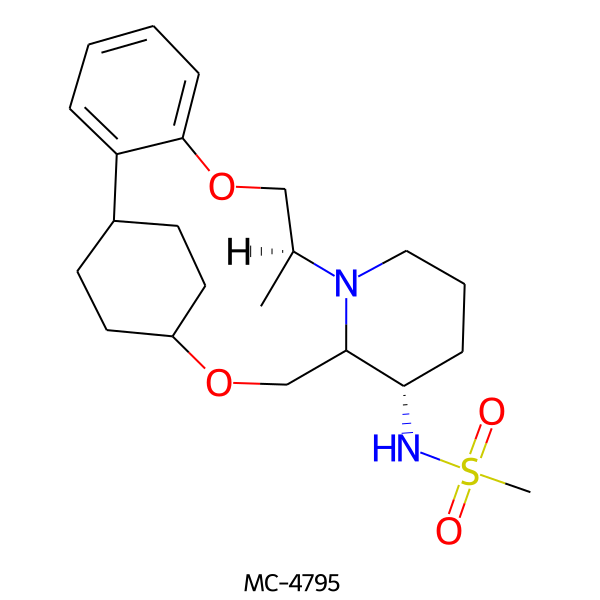 |
422.59 | 5 | 1 | 2.89 | 76.25 | 6.49 | |
| MC-4796 | 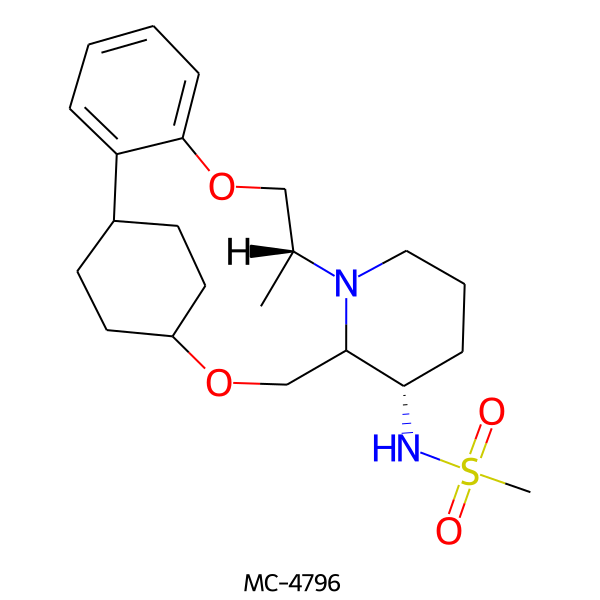 |
422.59 | 5 | 1 | 2.89 | 76.25 | 6.49 | |
| MC-4797 | 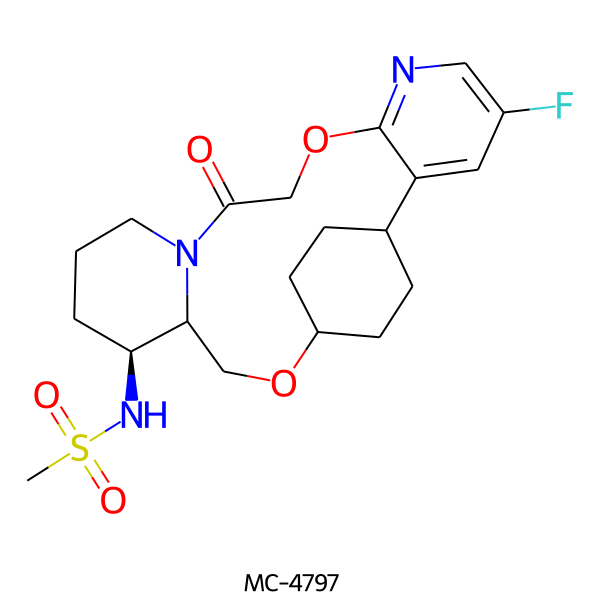 |
441.52 | 6 | 1 | 1.56 | 106.21 | 6.26 | |
| MC-4798 | 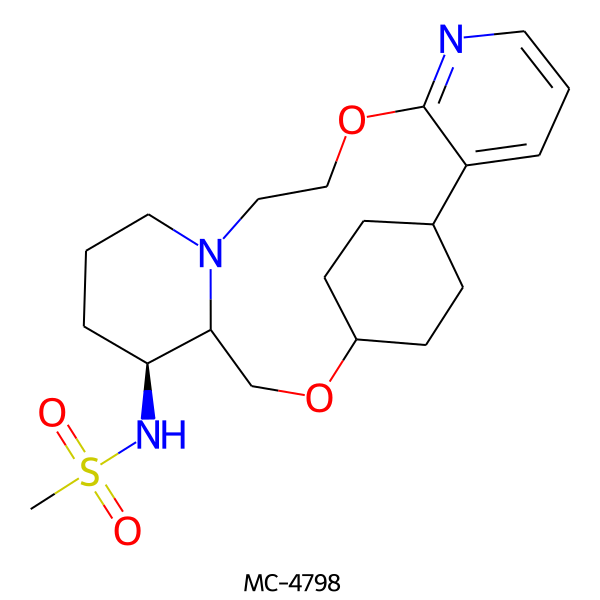 |
409.55 | 6 | 1 | 1.90 | 89.14 | 6.20 | |
| MC-4799 | 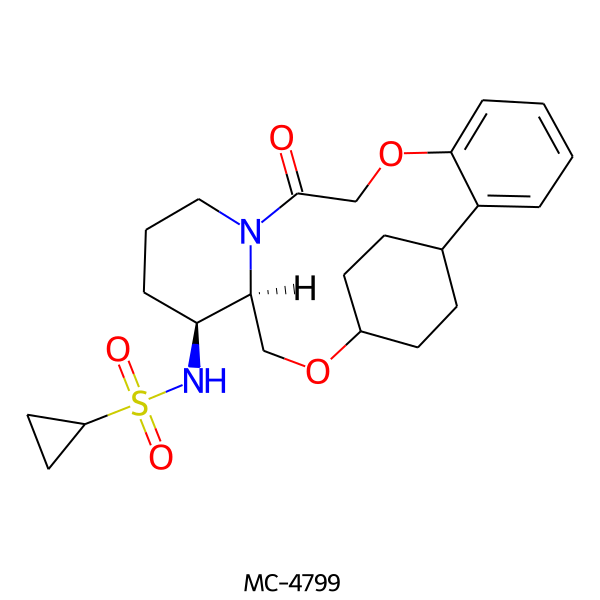 |
448.58 | 5 | 1 | 2.56 | 93.32 | 5.96 | |
| MC-4800 | 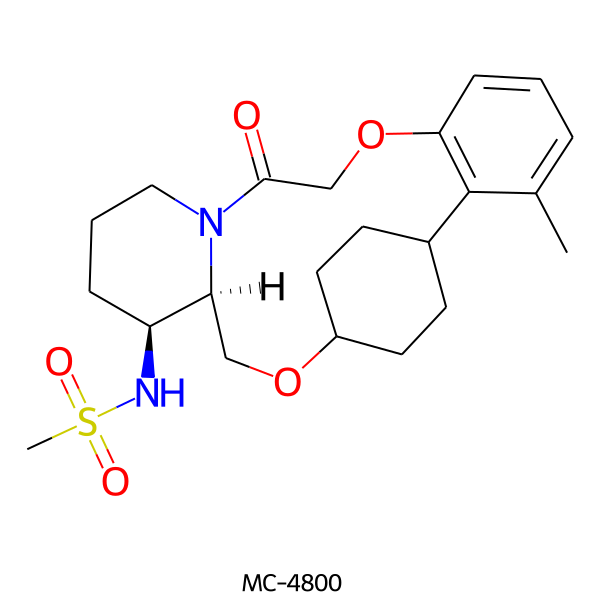 |
436.57 | 5 | 1 | 2.34 | 93.32 | 6.36 | |
| MC-4801 | 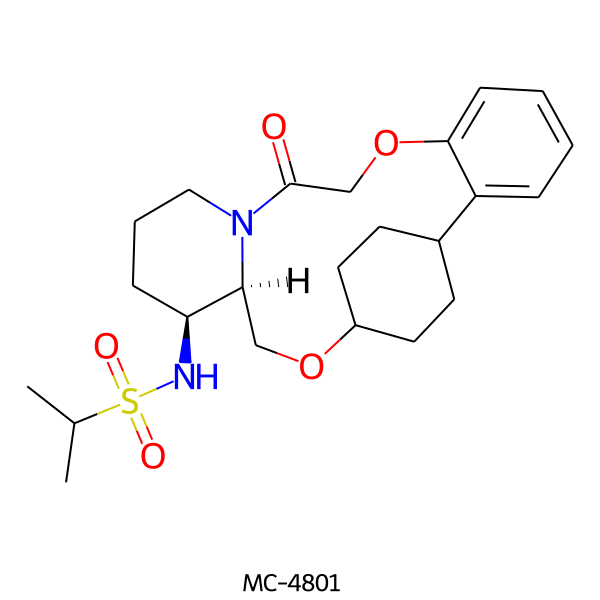 |
450.60 | 5 | 1 | 2.81 | 93.32 | 6.89 | |
| MC-4802 | 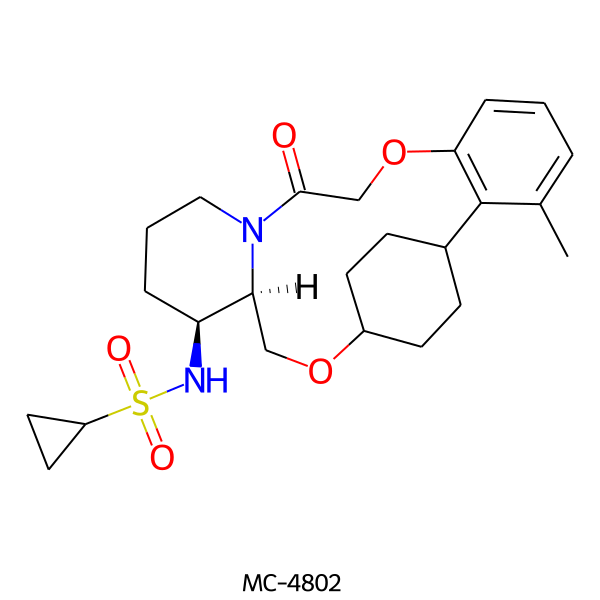 |
462.61 | 5 | 1 | 2.87 | 93.32 | 6.20 | |
| MC-4803 | 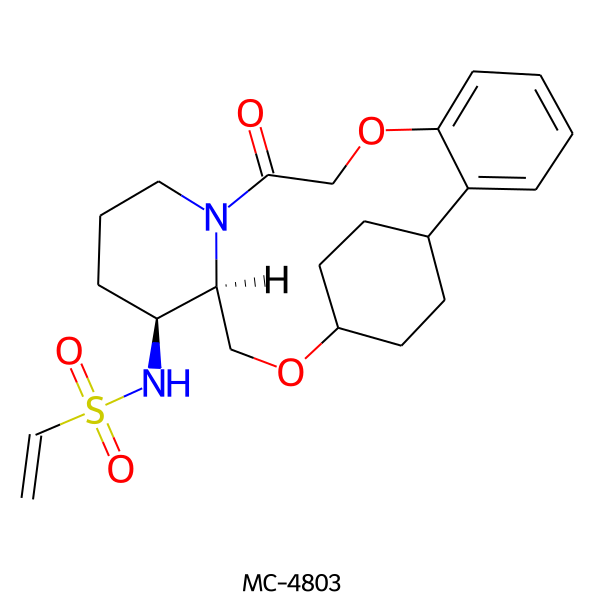 |
434.56 | 5 | 1 | 2.54 | 93.32 | 6.46 | |
| MC-4804 | 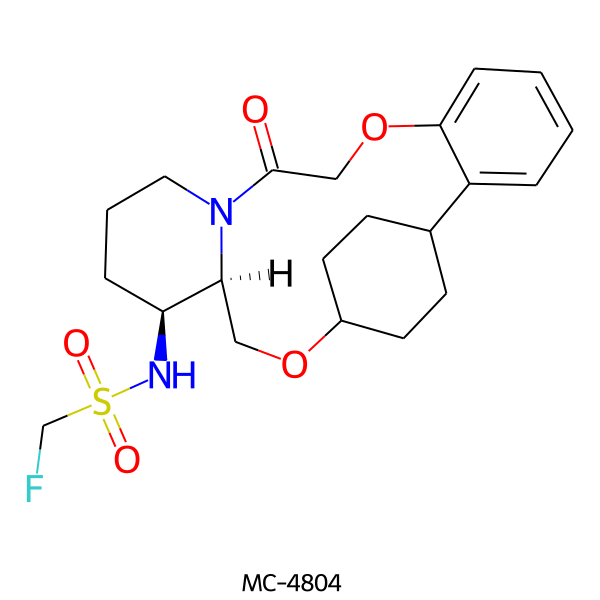 |
440.54 | 5 | 1 | 2.33 | 93.32 | 6.60 | |
| MC-4805 | 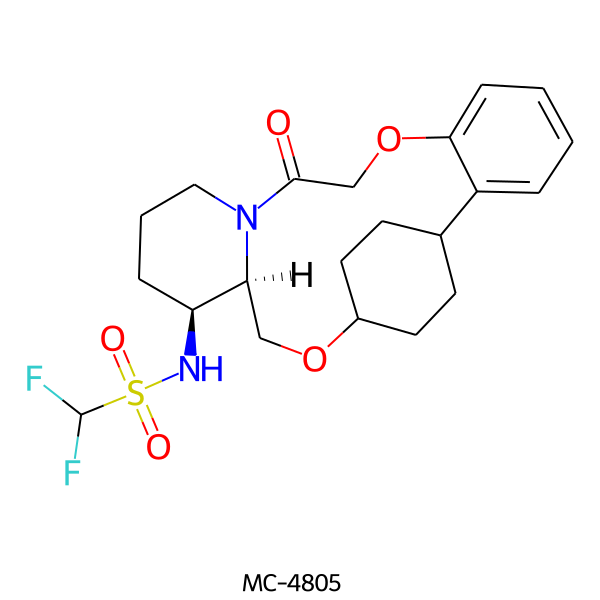 |
458.53 | 5 | 1 | 2.62 | 93.32 | 6.78 | |
| MC-4806 | 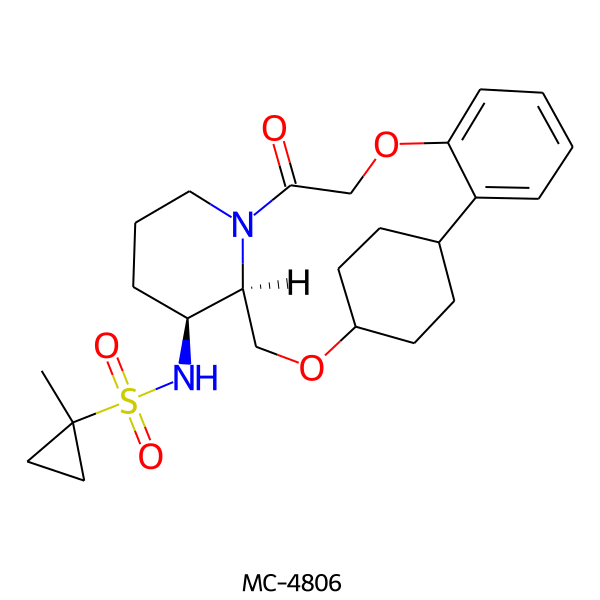 |
462.61 | 5 | 1 | 2.95 | 93.32 | 5.97 | |
| MC-4807 | 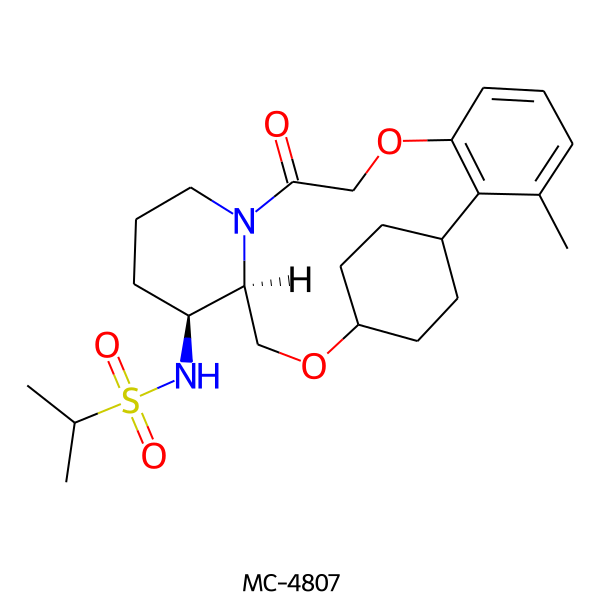 |
464.63 | 5 | 1 | 3.12 | 93.32 | 7.12 | |
| MC-4808 | 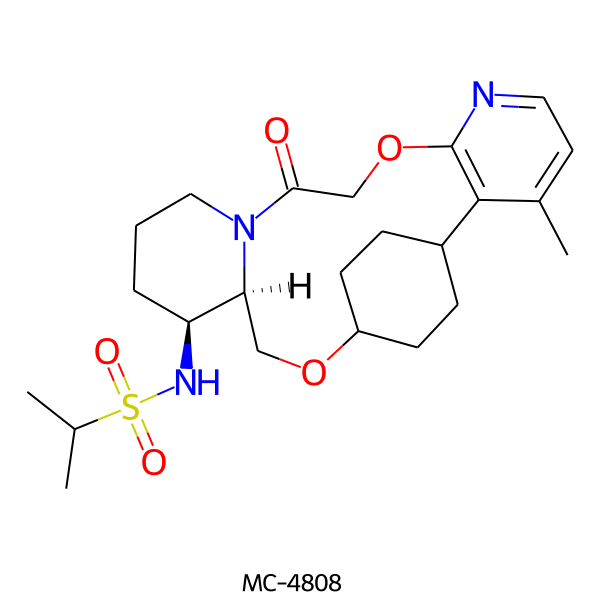 |
465.62 | 6 | 1 | 2.51 | 106.21 | 7.07 | |
| MC-4809 | 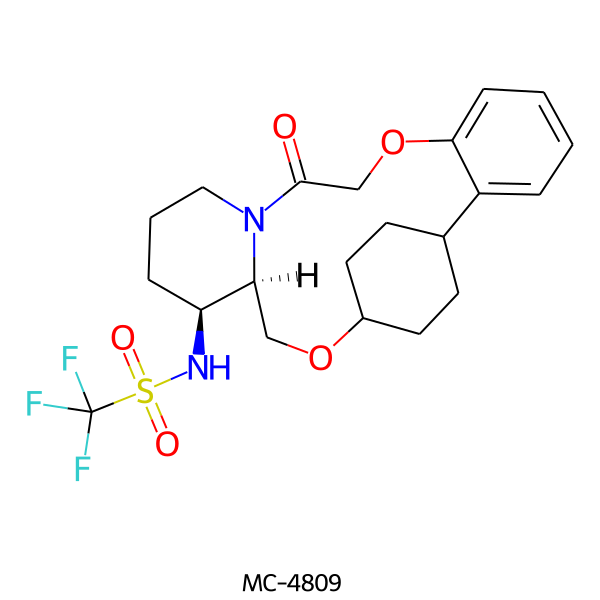 |
476.52 | 5 | 1 | 2.92 | 93.32 | 6.69 | |
| MC-4810 | 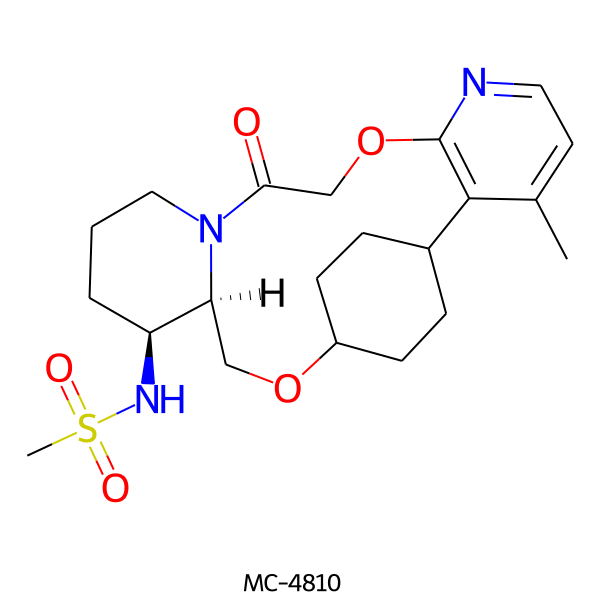 |
437.56 | 6 | 1 | 1.73 | 106.21 | 6.31 | |
| MC-4811 | 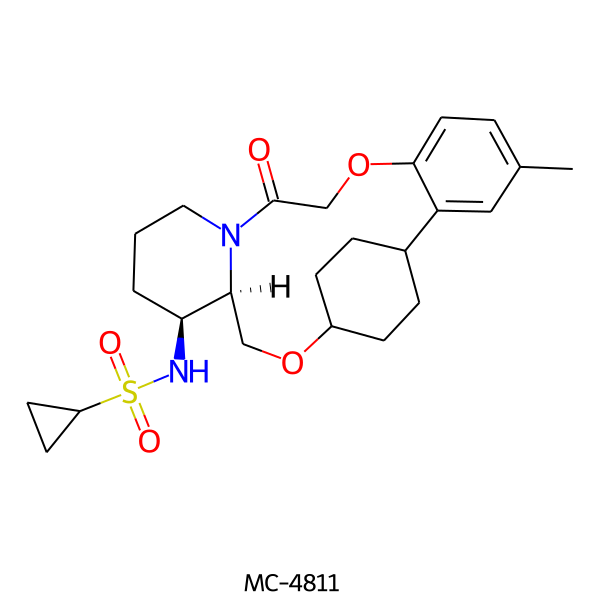 |
462.61 | 5 | 1 | 2.87 | 93.32 | 6.20 | |
| MC-4812 | 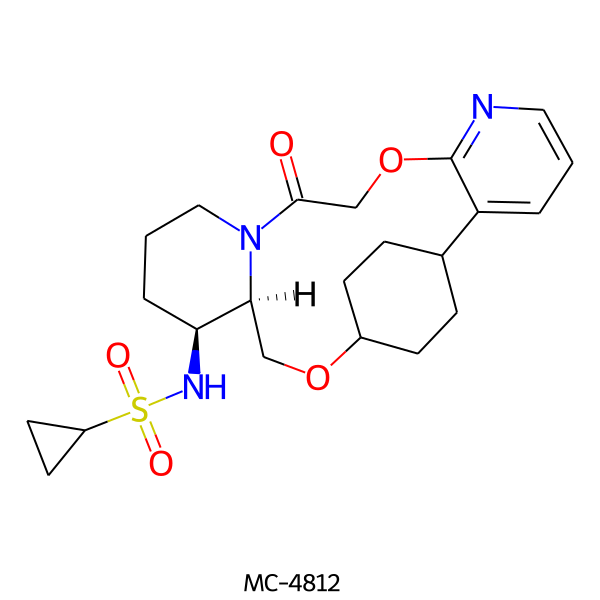 |
449.57 | 6 | 1 | 1.96 | 106.21 | 5.92 | |
| MC-4813 | 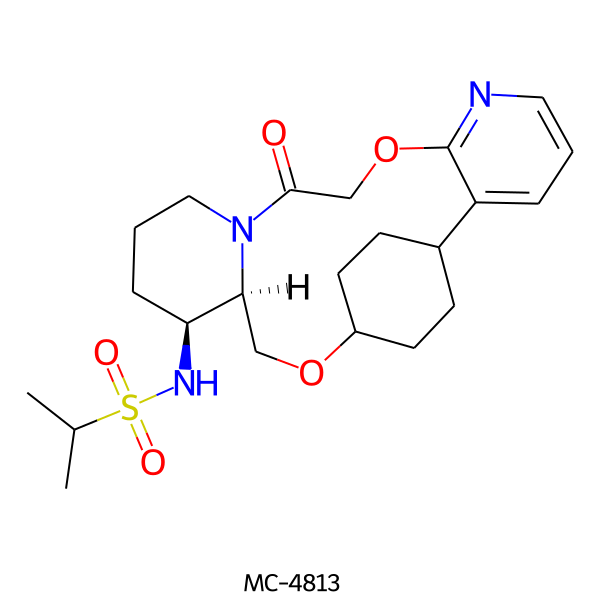 |
451.59 | 6 | 1 | 2.20 | 106.21 | 6.83 | |
| MC-4814 | 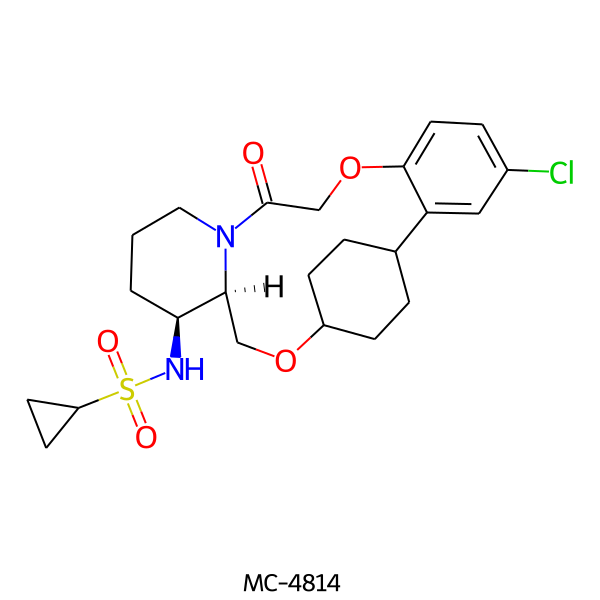 |
483.03 | 5 | 1 | 3.22 | 93.32 | 6.40 | |
| MC-4815 | 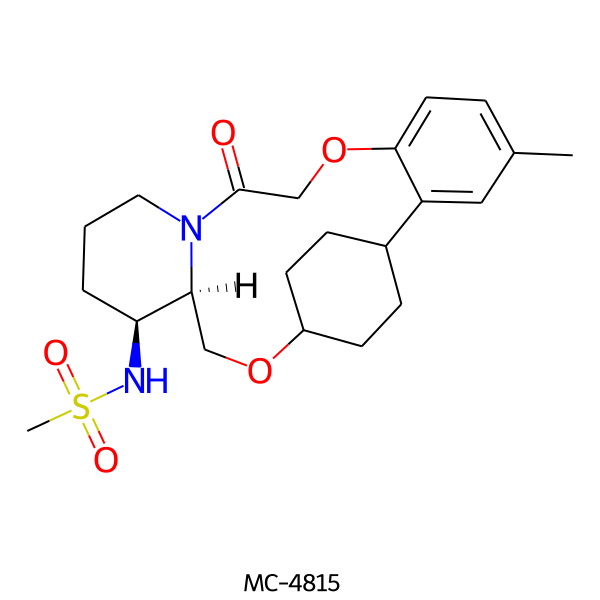 |
436.57 | 5 | 1 | 2.34 | 93.32 | 6.36 | |
| MC-4816 | 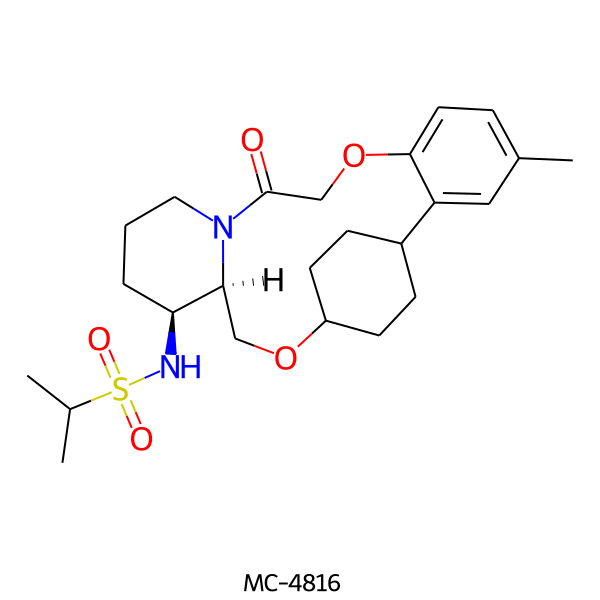 |
464.63 | 5 | 1 | 3.12 | 93.32 | 7.12 | |
| MC-4817 | 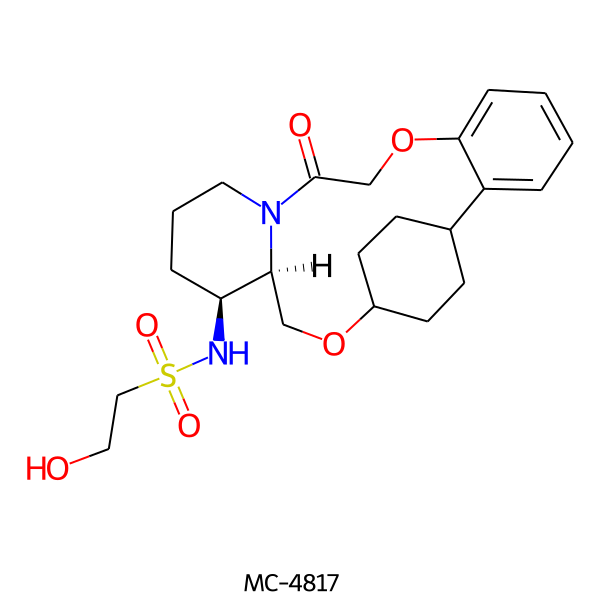 |
452.57 | 6 | 2 | 1.39 | 113.55 | 7.15 | |
| MC-4818 | 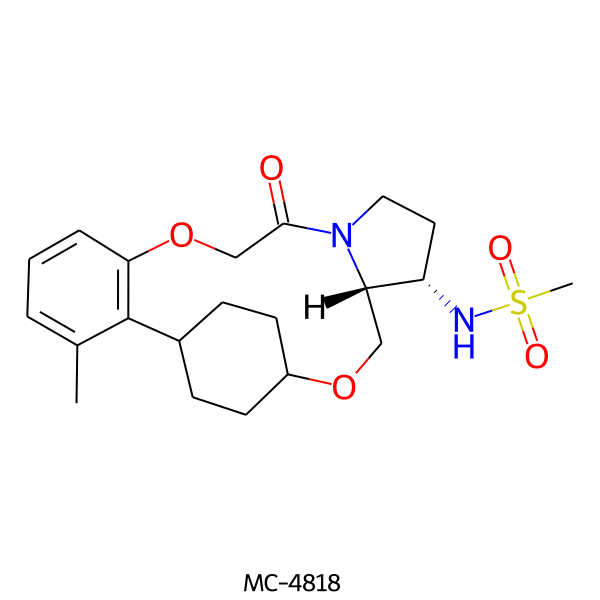 |
422.55 | 5 | 1 | 1.95 | 93.32 | 5.86 | |
| MC-4819 | 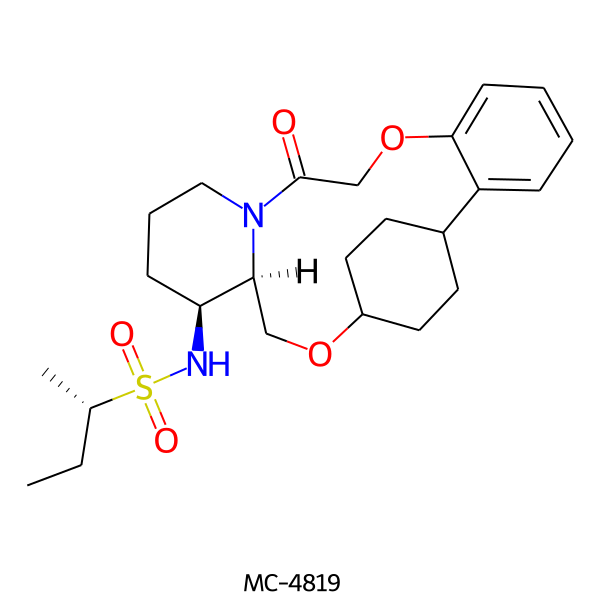 |
464.63 | 5 | 1 | 3.20 | 93.32 | 7.42 | |
| MC-4820 | 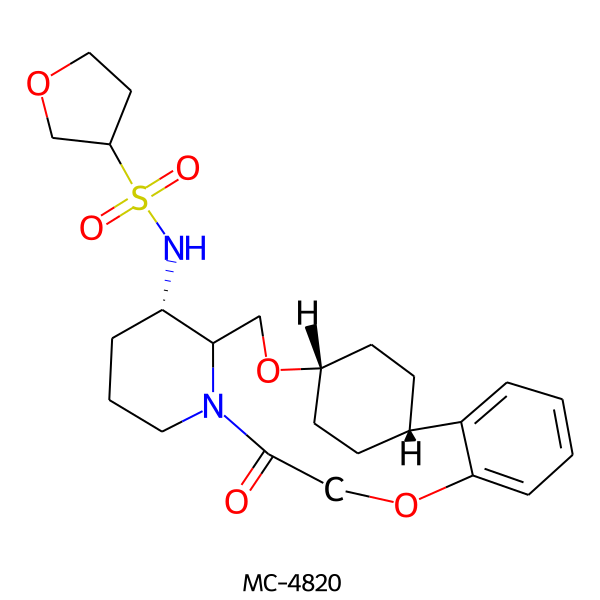 |
478.61 | 6 | 1 | 2.19 | 102.55 | 6.92 | |
| MC-4821 | 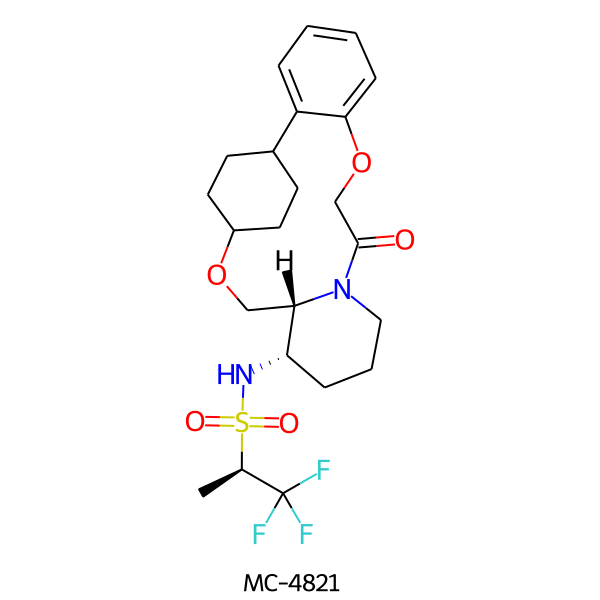 |
504.57 | 5 | 1 | 3.35 | 93.32 | 7.44 | |
| MC-4822 | 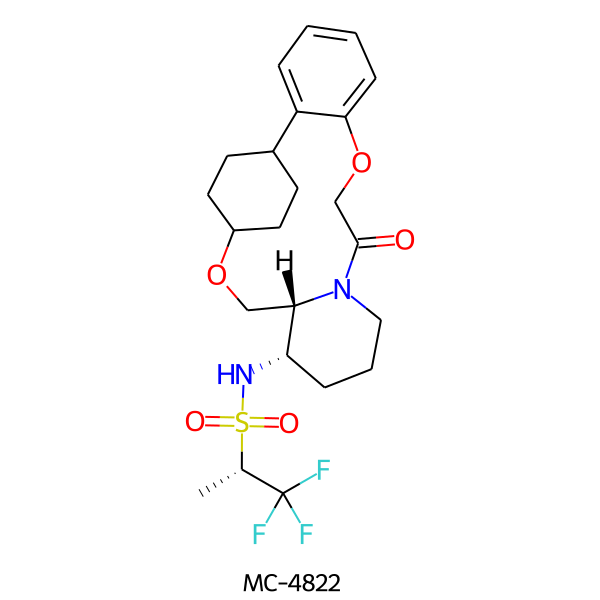 |
504.57 | 5 | 1 | 3.35 | 93.32 | 7.44 | |
| MC-4823 | 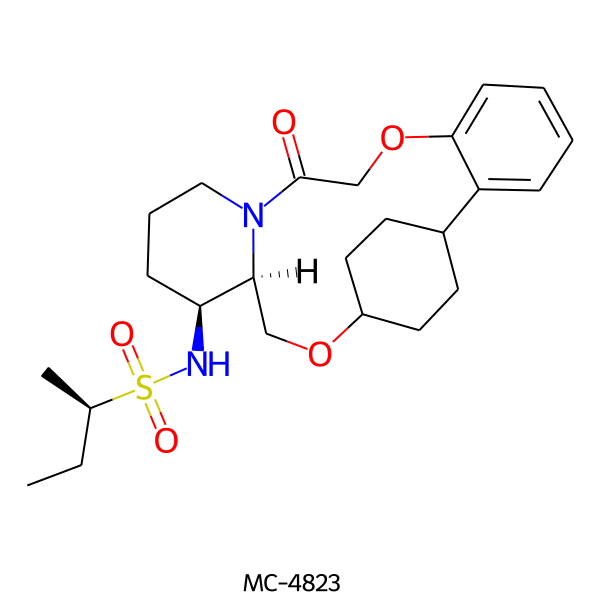 |
464.63 | 5 | 1 | 3.20 | 93.32 | 7.42 | |
| MC-4824 | 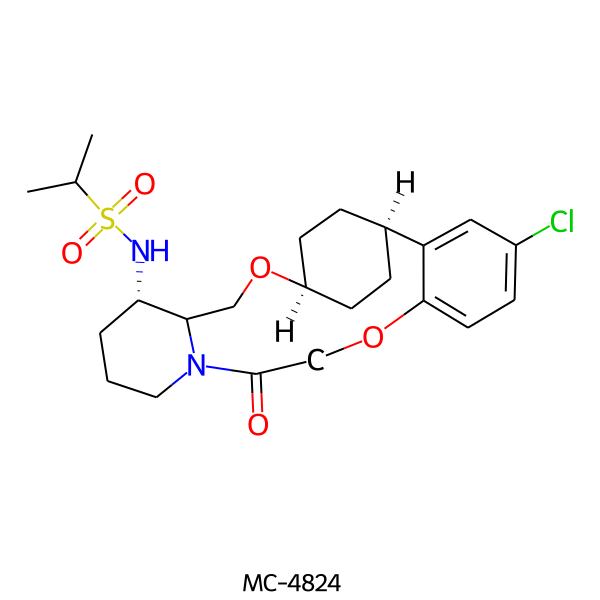 |
485.05 | 5 | 1 | 3.46 | 93.32 | 7.34 | |
| MC-4825 | 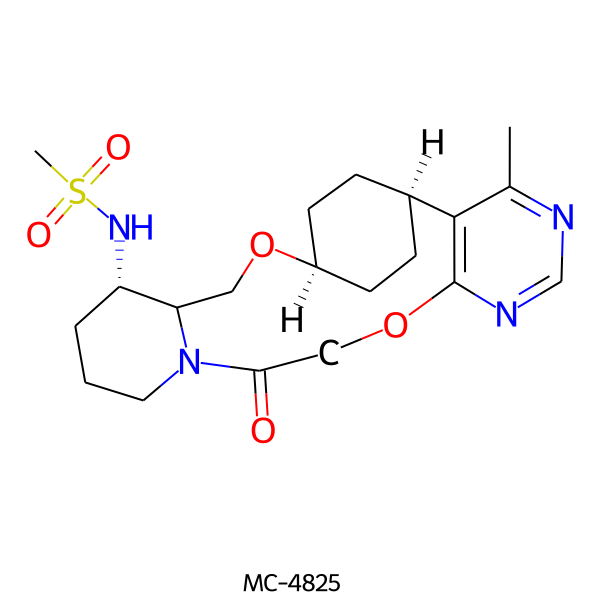 |
438.55 | 7 | 1 | 1.13 | 119.10 | 6.26 | |
| MC-4826 | 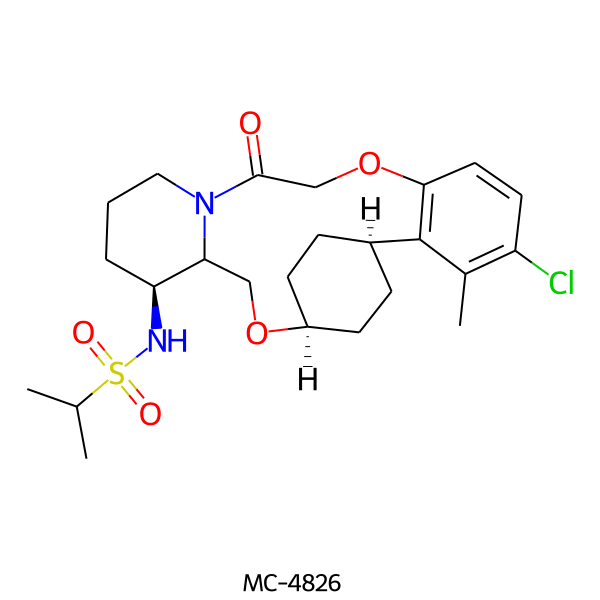 |
499.07 | 5 | 1 | 3.77 | 93.32 | 7.58 | |
| MC-4827 | 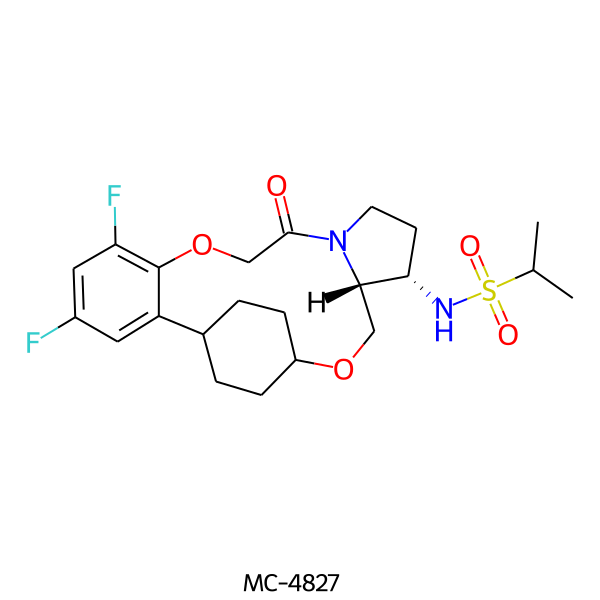 |
472.55 | 5 | 1 | 2.70 | 93.32 | 6.74 | |
| MC-4828 | 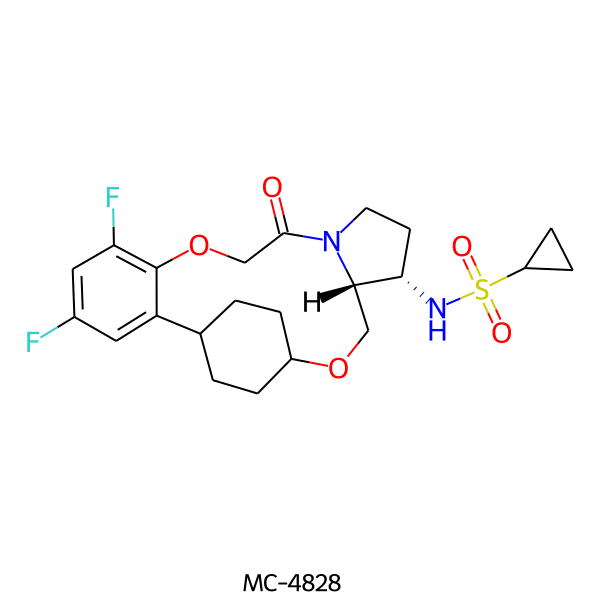 |
470.54 | 5 | 1 | 2.45 | 93.32 | 5.88 | |
| MC-4829 | 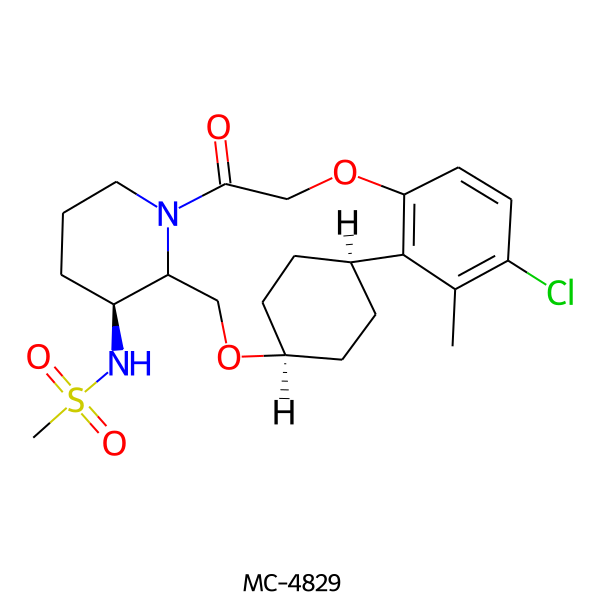 |
471.02 | 5 | 1 | 2.99 | 93.32 | 6.82 | |
| MC-4830 | 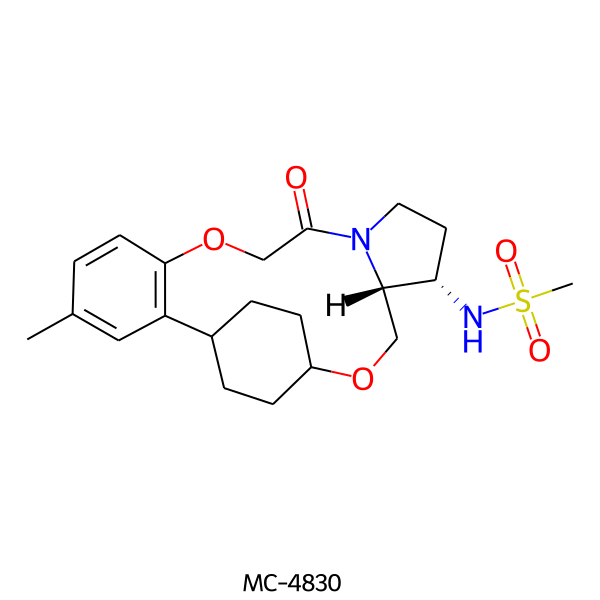 |
422.55 | 5 | 1 | 1.95 | 93.32 | 5.86 | |
| MC-4831 | 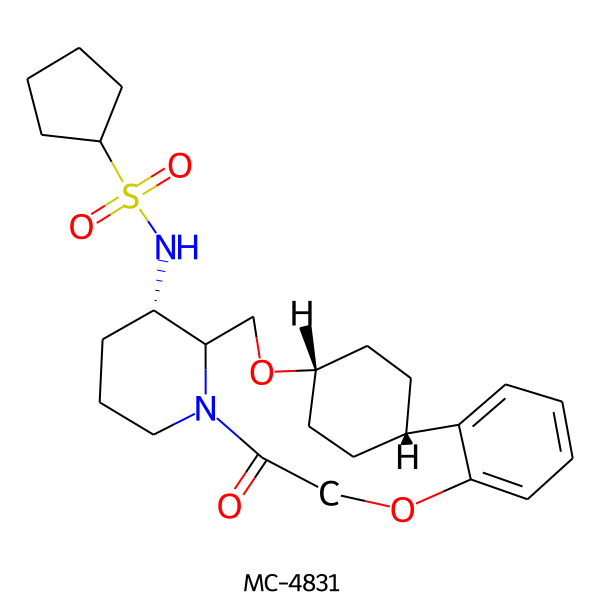 |
476.64 | 5 | 1 | 3.34 | 93.32 | 6.95 | |
| MC-4832 | 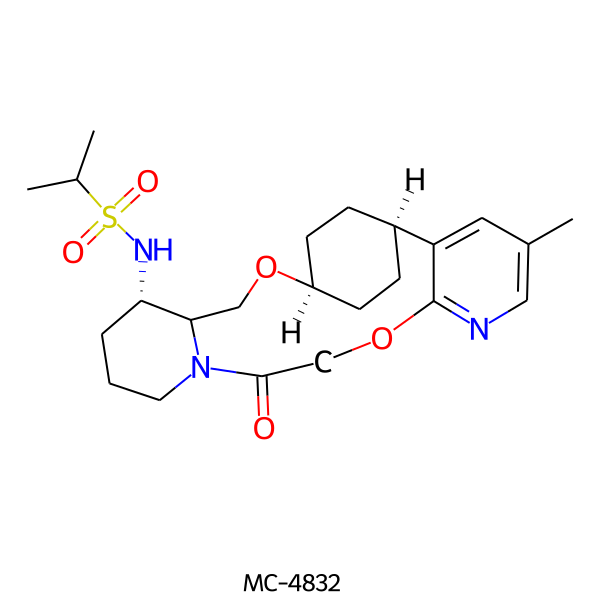 |
465.62 | 6 | 1 | 2.51 | 106.21 | 7.07 | |
| MC-4833 | 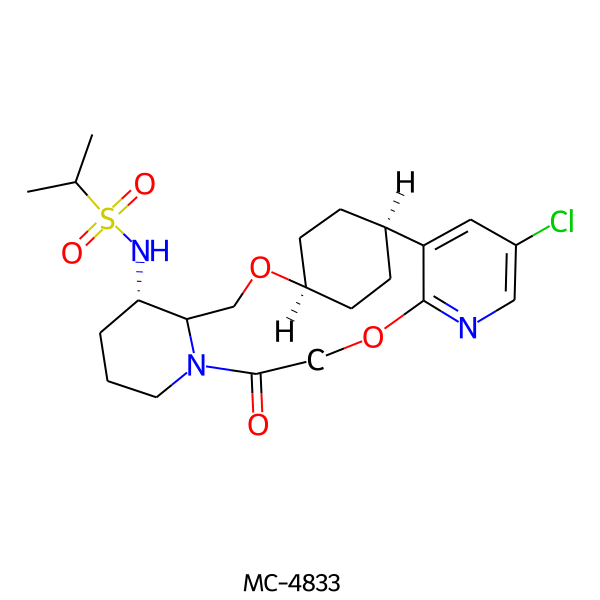 |
486.03 | 6 | 1 | 2.86 | 106.21 | 7.29 | |
| MC-4834 | 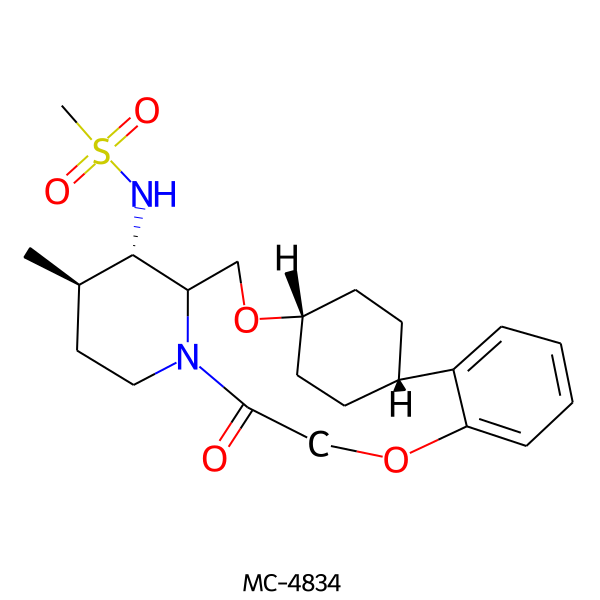 |
436.57 | 5 | 1 | 2.28 | 93.32 | 6.36 | |
| MC-4835 | 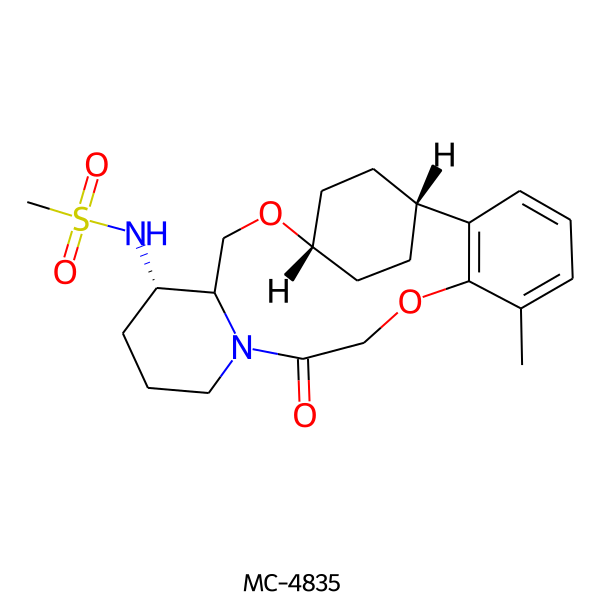 |
436.57 | 5 | 1 | 2.34 | 93.32 | 6.36 | |
| MC-4836 | 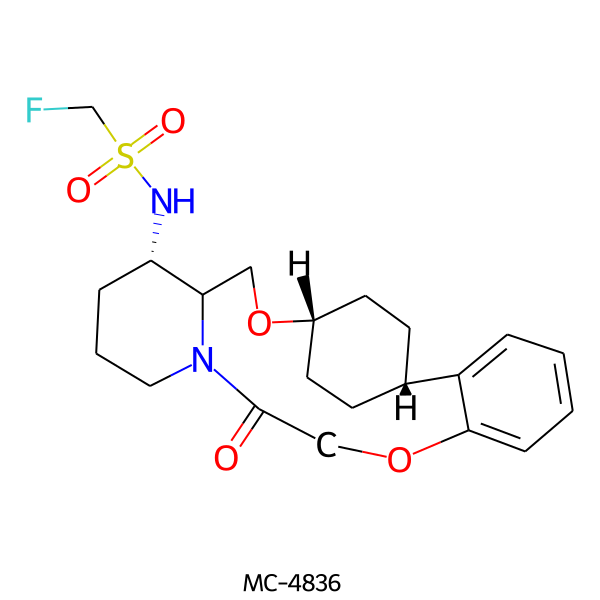 |
440.54 | 5 | 1 | 2.33 | 93.32 | 6.60 | |
| MC-4837 | 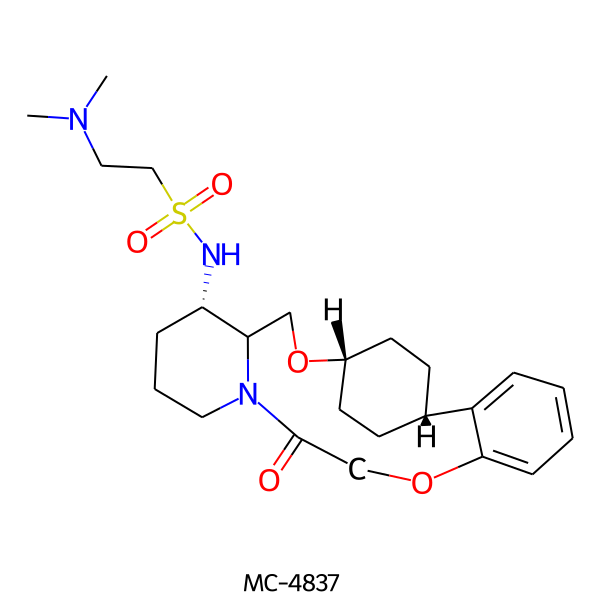 |
479.64 | 6 | 1 | 1.96 | 96.56 | 7.94 | |
| MC-4838 | 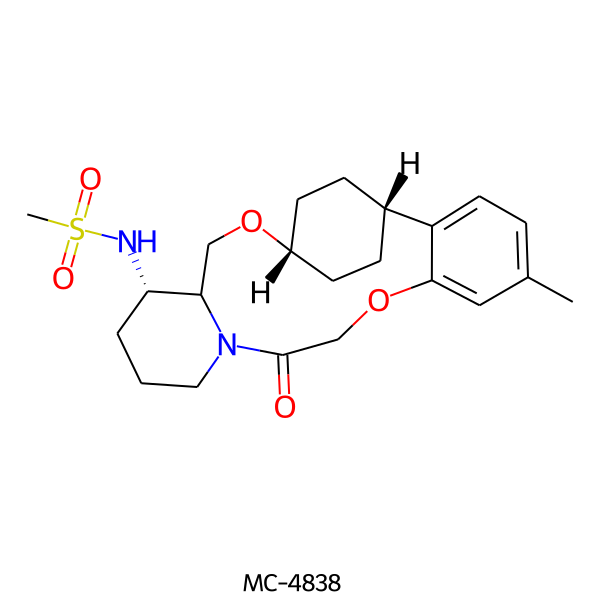 |
436.57 | 5 | 1 | 2.34 | 93.32 | 6.36 | |
| MC-4839 | 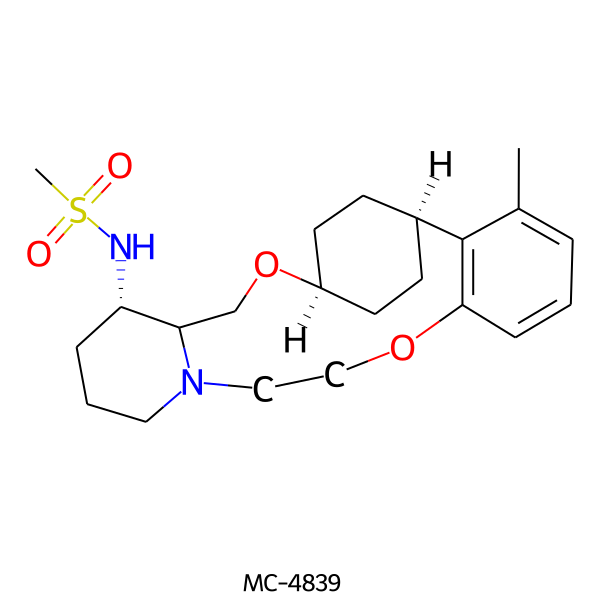 |
422.59 | 5 | 1 | 2.81 | 76.25 | 6.49 | |
| MC-4840 | 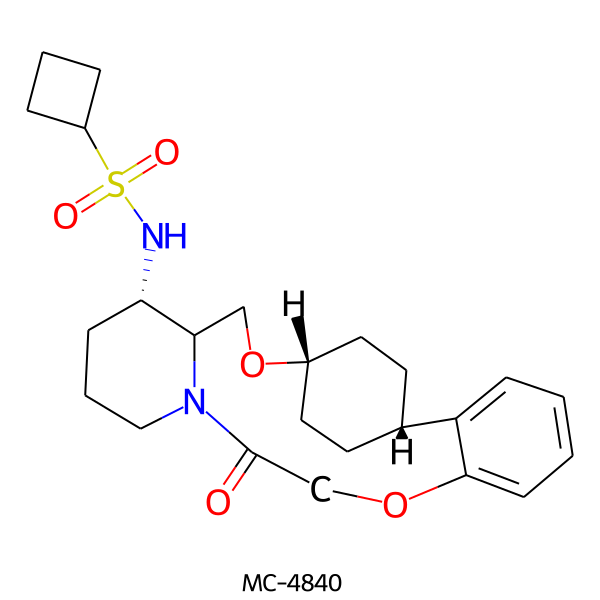 |
462.61 | 5 | 1 | 2.95 | 93.32 | 6.45 | |
| MC-4841 | 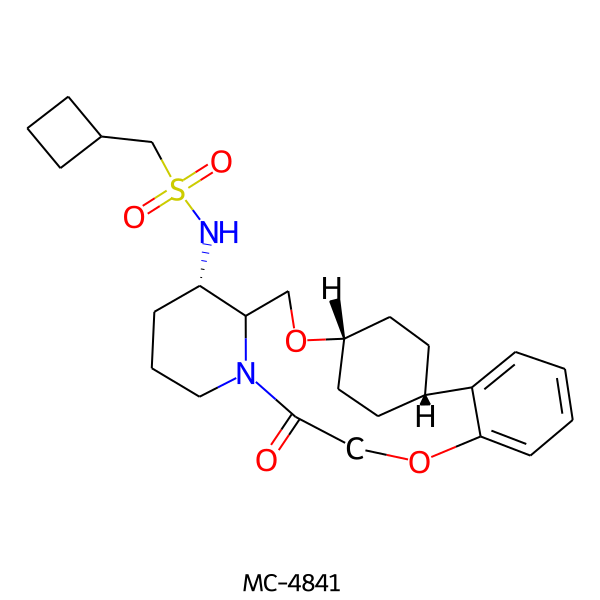 |
476.64 | 5 | 1 | 3.20 | 93.32 | 6.95 | |
| MC-4842 | 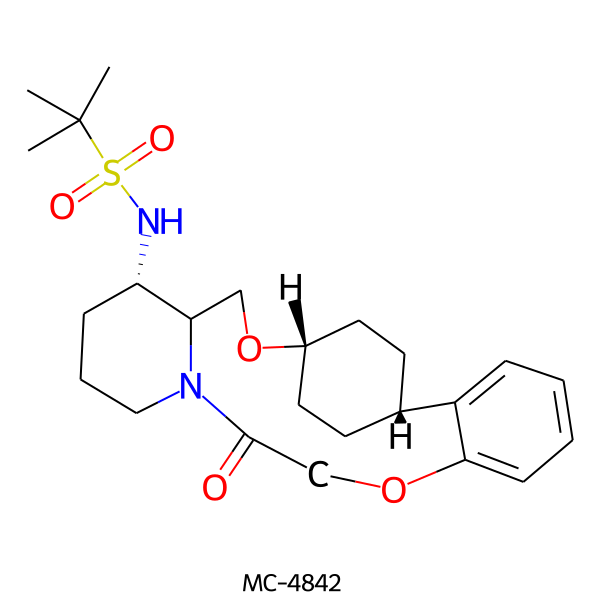 |
464.63 | 5 | 1 | 3.20 | 93.32 | 6.84 | |
| MC-4843 | 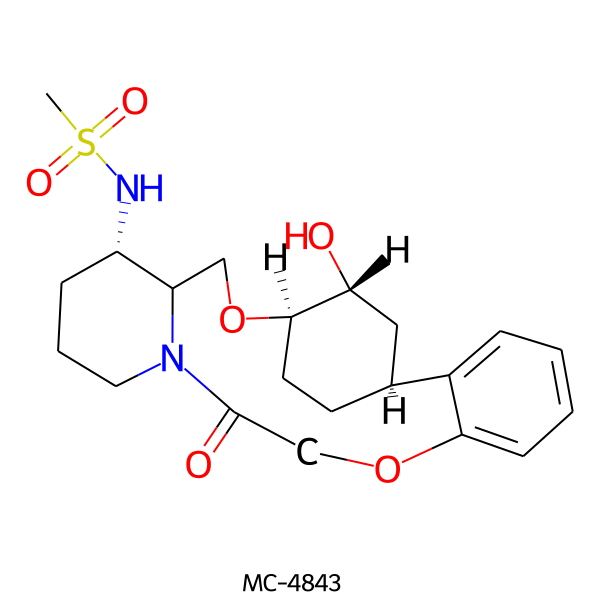 |
438.55 | 6 | 2 | 1.00 | 113.55 | 6.34 | |
| MC-4844 | 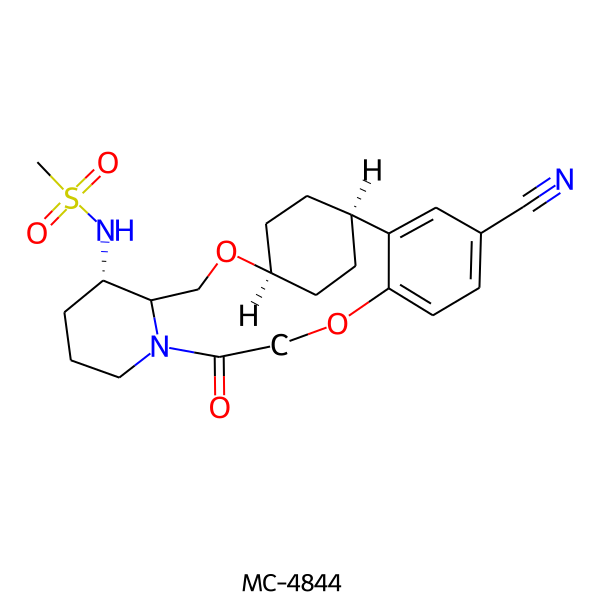 |
447.56 | 6 | 1 | 1.90 | 117.11 | 6.51 | |
| MC-4845 | 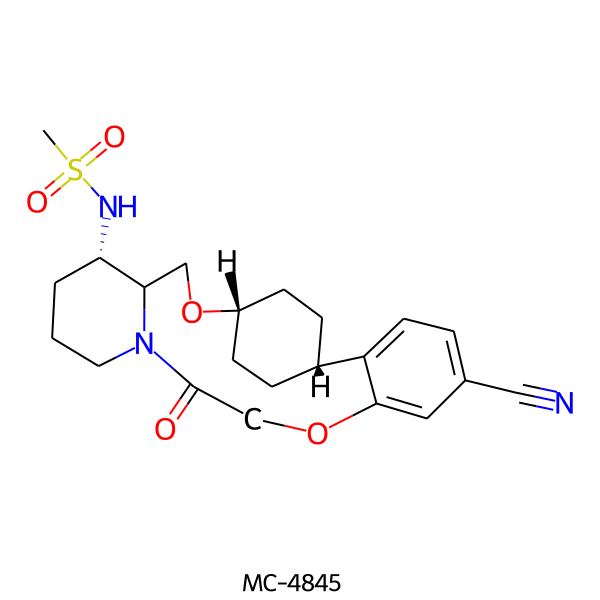 |
447.56 | 6 | 1 | 1.90 | 117.11 | 6.51 | |
| MC-4846 | 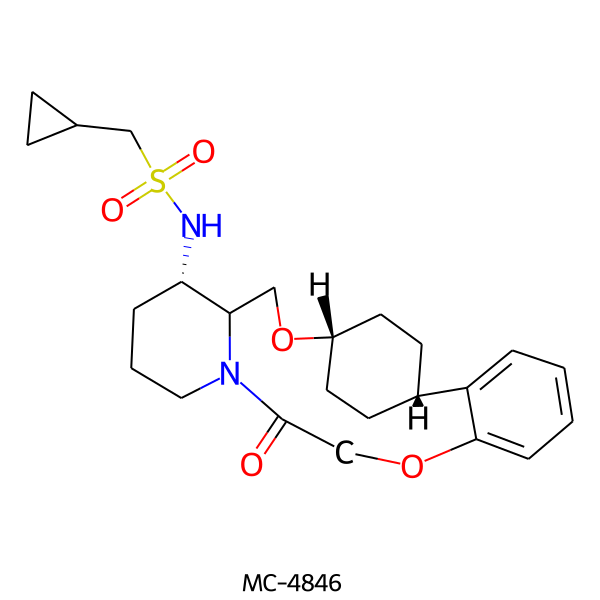 |
462.61 | 5 | 1 | 2.81 | 93.32 | 6.45 | |
| MC-4847 | 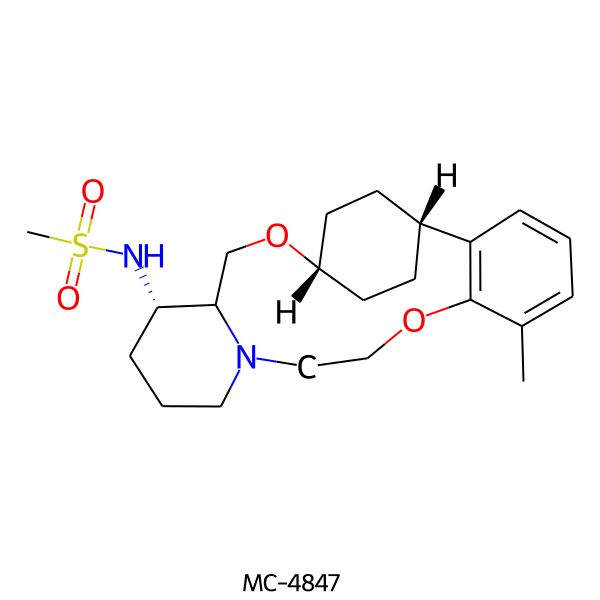 |
422.59 | 5 | 1 | 2.81 | 76.25 | 6.49 | |
| MC-4848 | 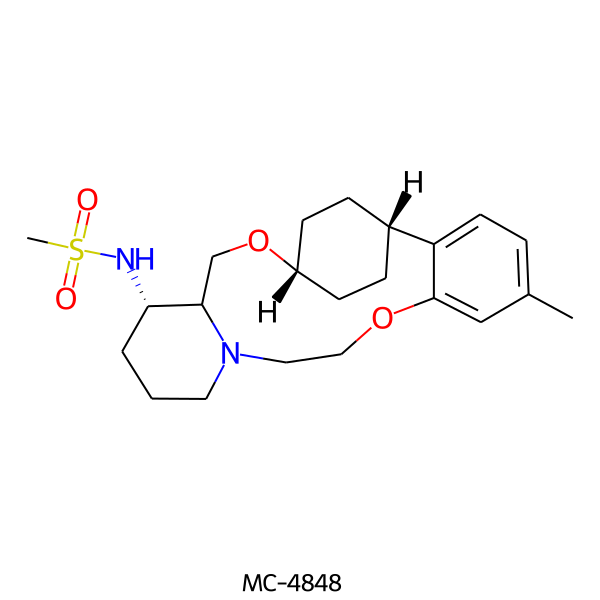 |
422.59 | 5 | 1 | 2.81 | 76.25 | 6.49 | |
| MC-4849 | 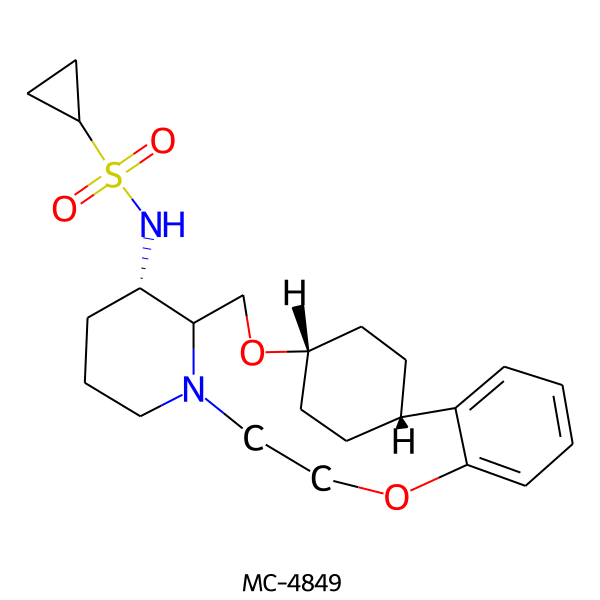 |
434.60 | 5 | 1 | 3.04 | 76.25 | 6.06 | |
| MC-4850 | 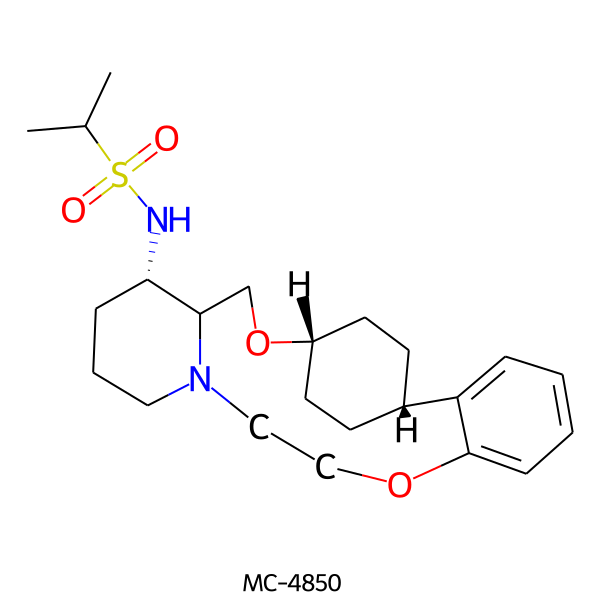 |
436.62 | 5 | 1 | 3.28 | 76.25 | 7.02 | |
| MC-4851 | 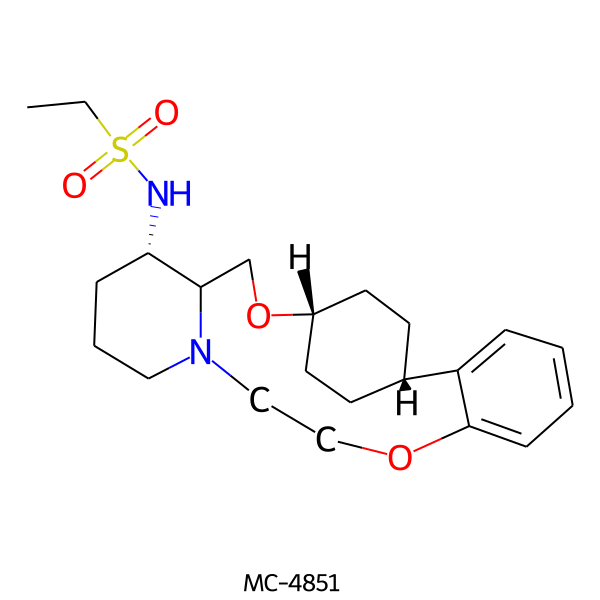 |
422.59 | 5 | 1 | 2.89 | 76.25 | 6.79 | |
| MC-4852 | 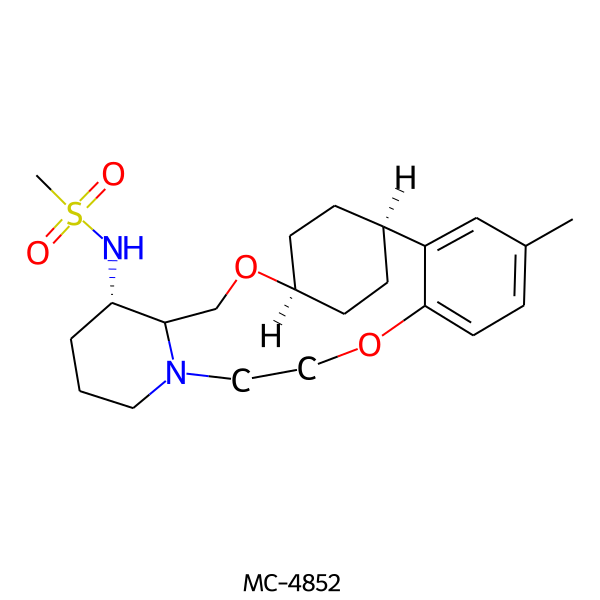 |
422.59 | 5 | 1 | 2.81 | 76.25 | 6.49 | |
| MC-4853 | 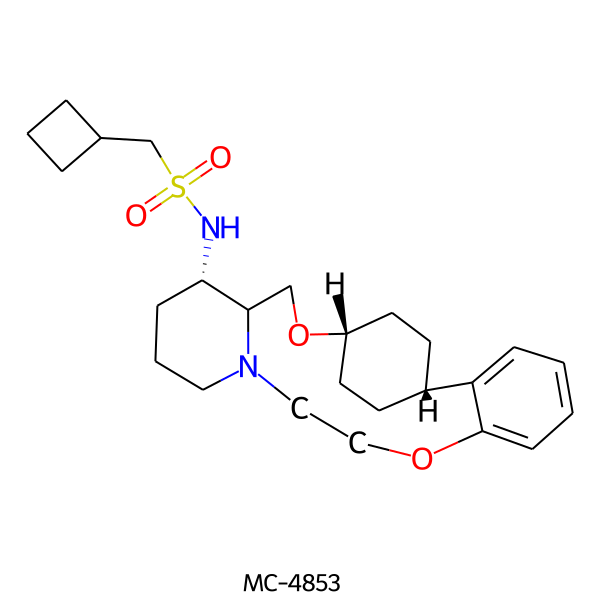 |
462.66 | 5 | 1 | 3.67 | 76.25 | 7.07 | |
| MC-4854 | 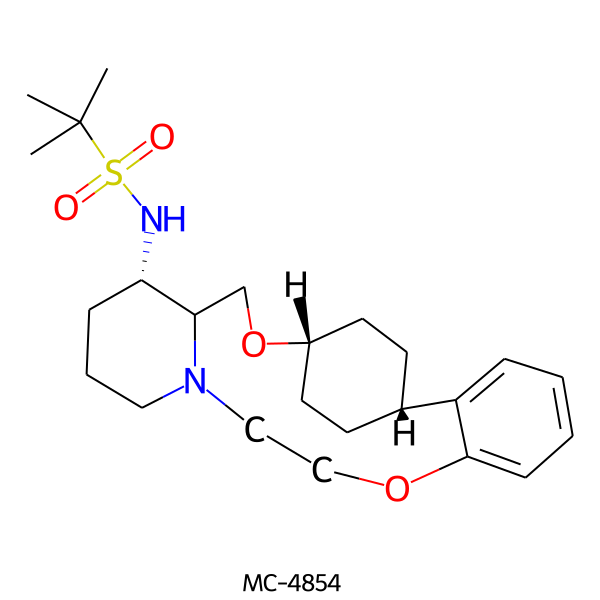 |
450.64 | 5 | 1 | 3.67 | 76.25 | 6.96 | |
| MC-4855 | 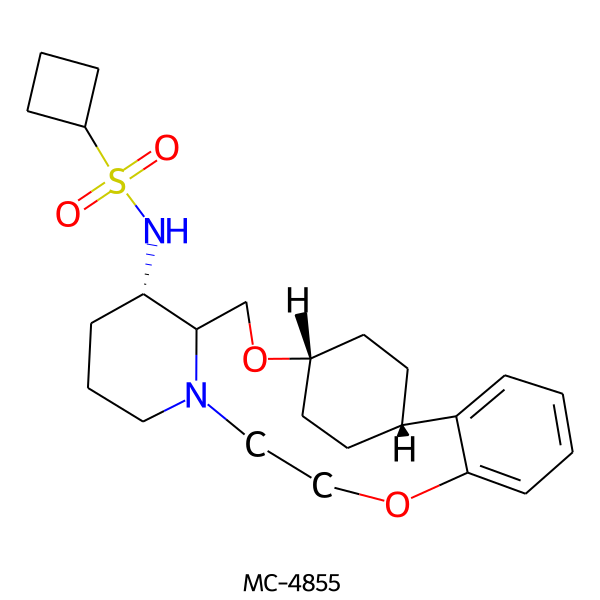 |
448.63 | 5 | 1 | 3.43 | 76.25 | 6.56 | |
| MC-4856 | 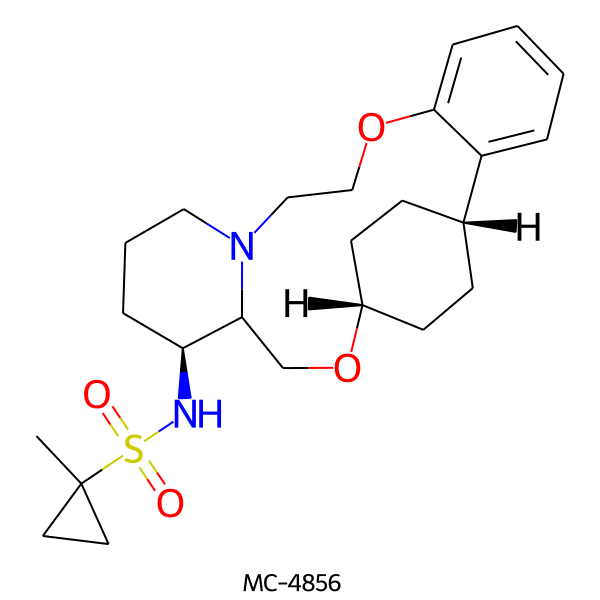 |
448.63 | 5 | 1 | 3.43 | 76.25 | 6.05 | |
| MC-4857 | 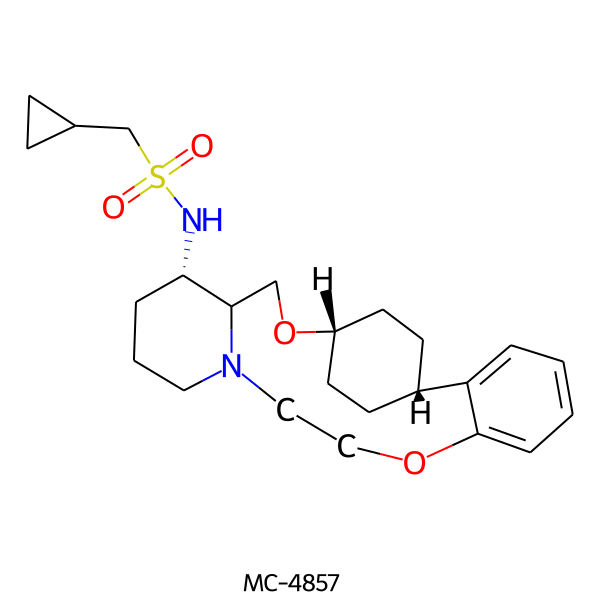 |
448.63 | 5 | 1 | 3.28 | 76.25 | 6.56 | |
| MC-4858 | 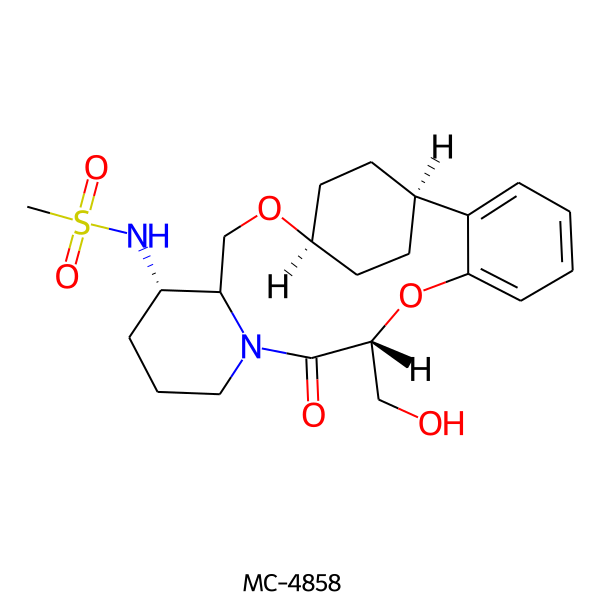 |
452.57 | 6 | 2 | 1.39 | 113.55 | 6.86 | |
| MC-4859 | 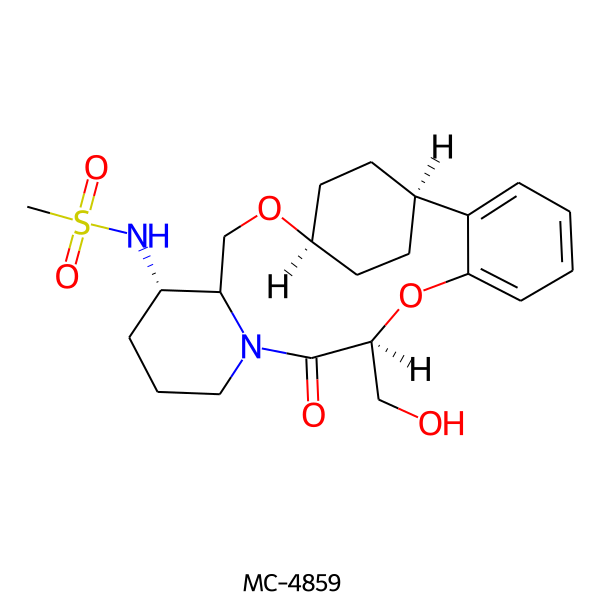 |
452.57 | 6 | 2 | 1.39 | 113.55 | 6.86 | |
| MC-4860 | 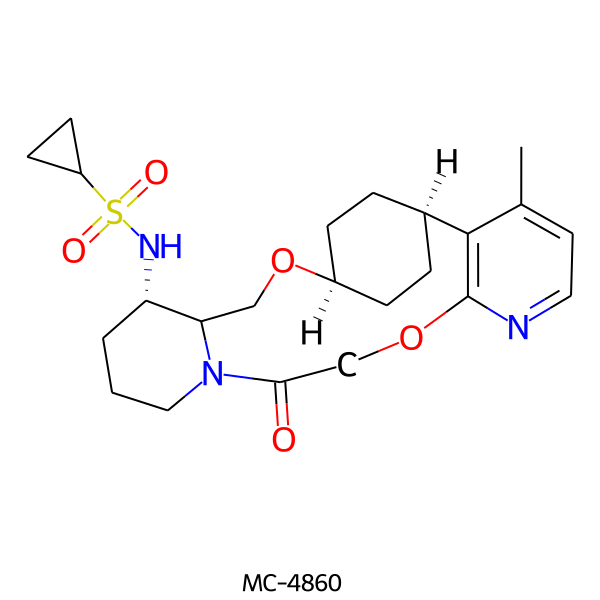 |
463.60 | 6 | 1 | 2.27 | 106.21 | 6.15 | |
| MC-4861 | 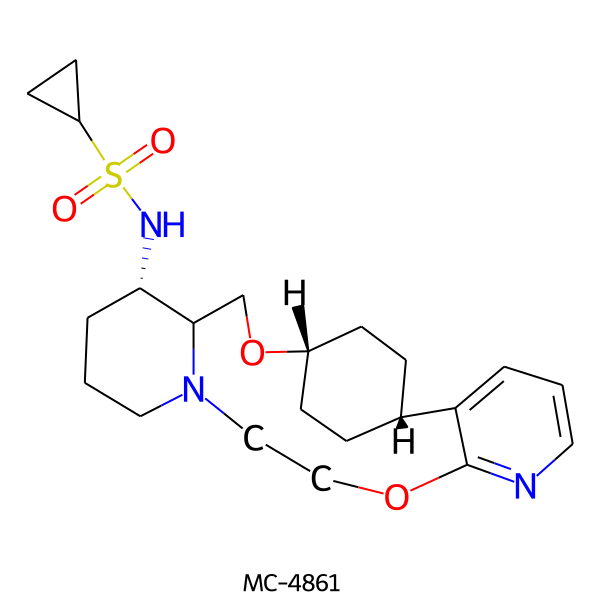 |
435.59 | 6 | 1 | 2.43 | 89.14 | 6.01 | |
| MC-4862 | 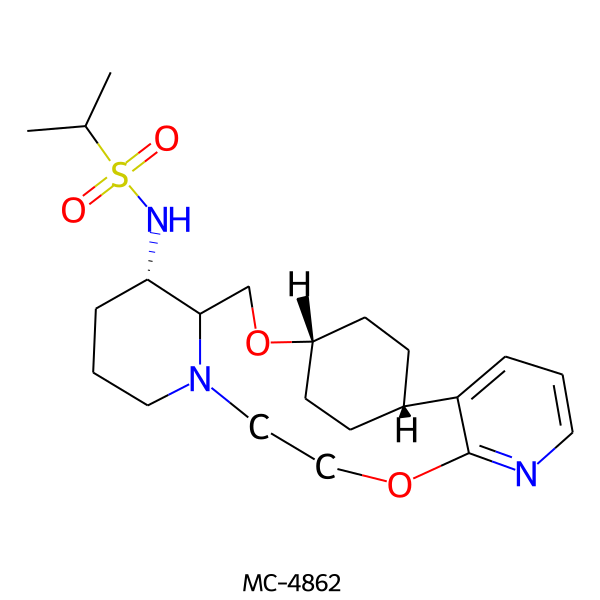 |
437.61 | 6 | 1 | 2.68 | 89.14 | 6.97 | |
| MC-4863 | 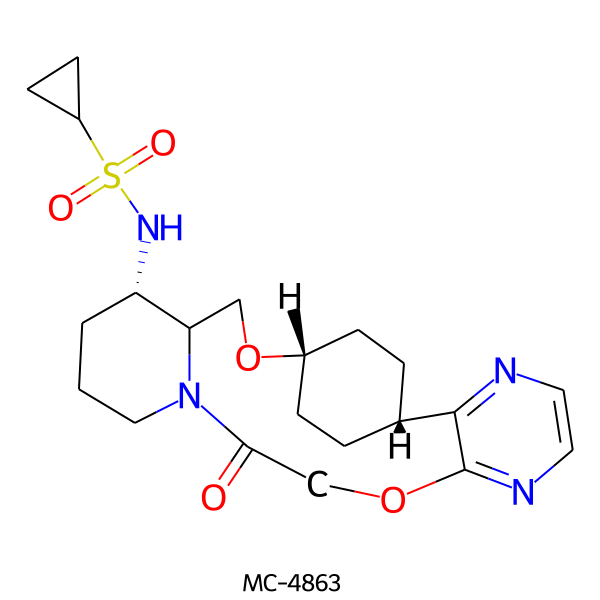 |
450.56 | 7 | 1 | 1.35 | 119.10 | 5.87 | |
| MC-4864 | 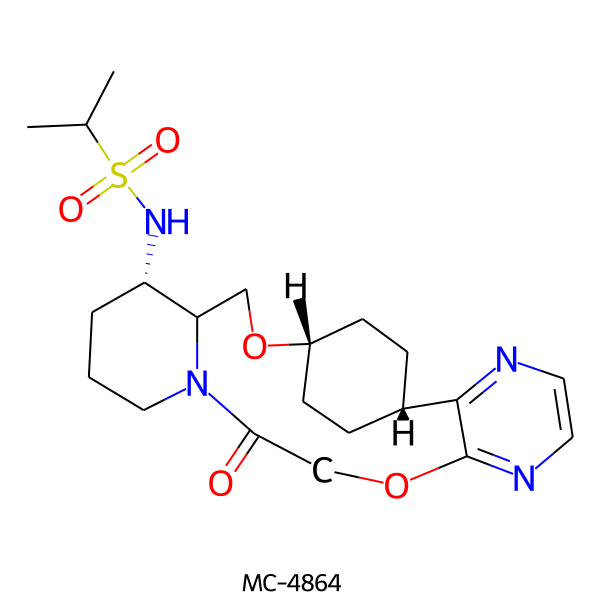 |
452.58 | 7 | 1 | 1.60 | 119.10 | 6.78 | |
| MC-4865 | 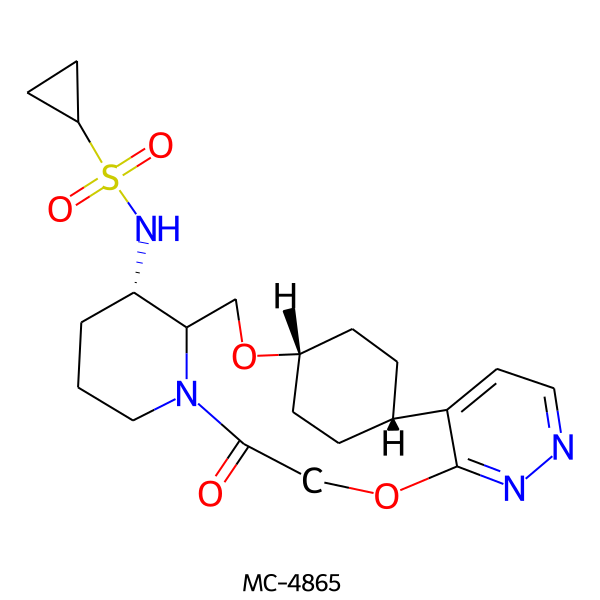 |
450.56 | 7 | 1 | 1.35 | 119.10 | 5.87 | |
| MC-4866 | 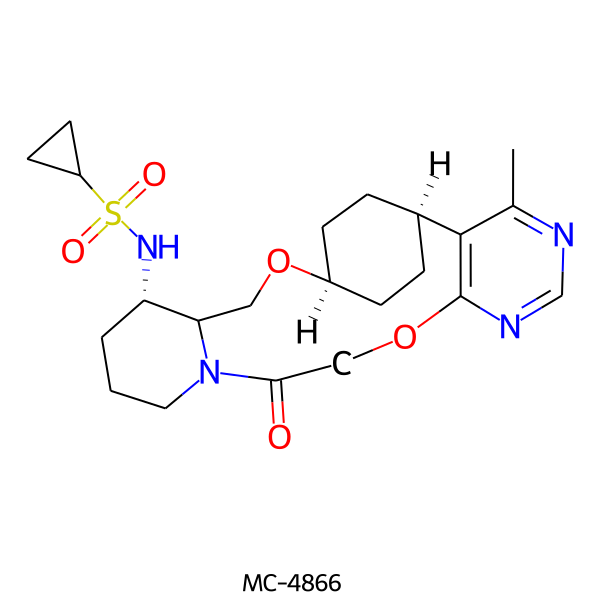 |
464.59 | 7 | 1 | 1.66 | 119.10 | 6.11 | |
| MC-4867 | 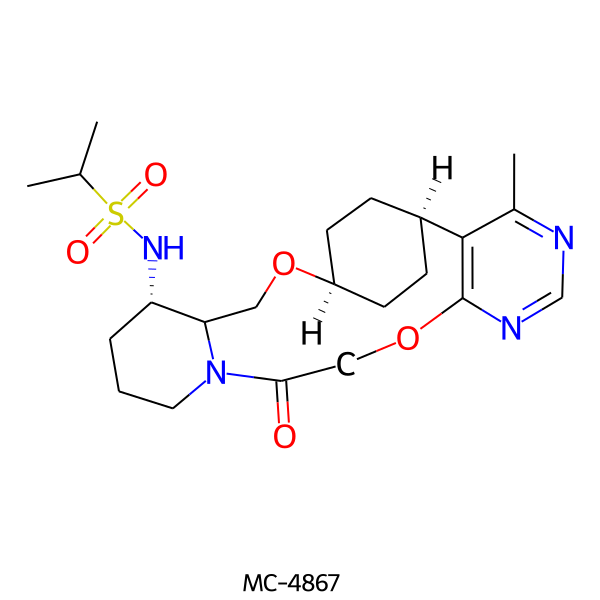 |
466.60 | 7 | 1 | 1.91 | 119.10 | 7.02 | |
| MC-4868 | 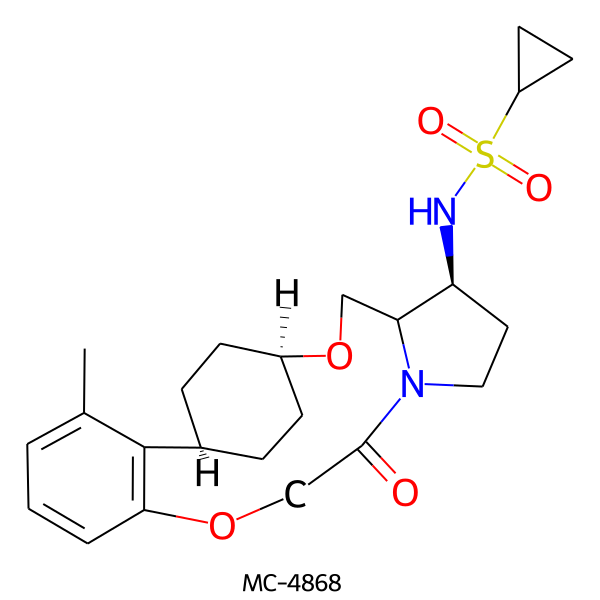 |
448.58 | 5 | 1 | 2.48 | 93.32 | 5.73 | |
| MC-4869 | 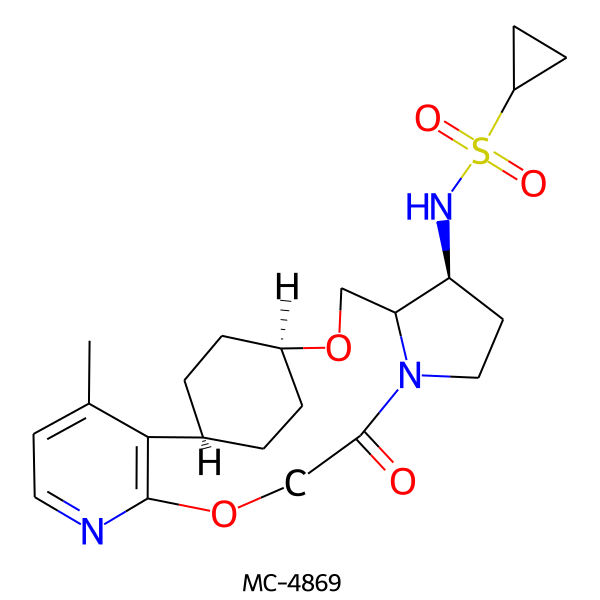 |
449.57 | 6 | 1 | 1.88 | 106.21 | 5.68 | |
| MC-4870 | 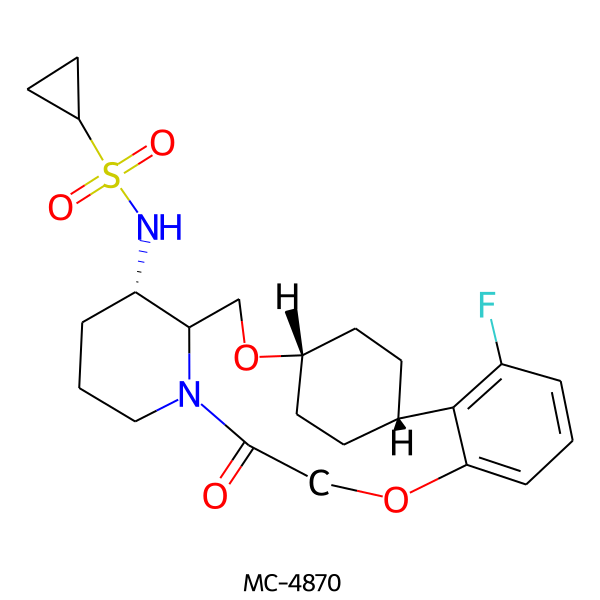 |
466.58 | 5 | 1 | 2.70 | 93.32 | 6.15 | |
| MC-4871 | 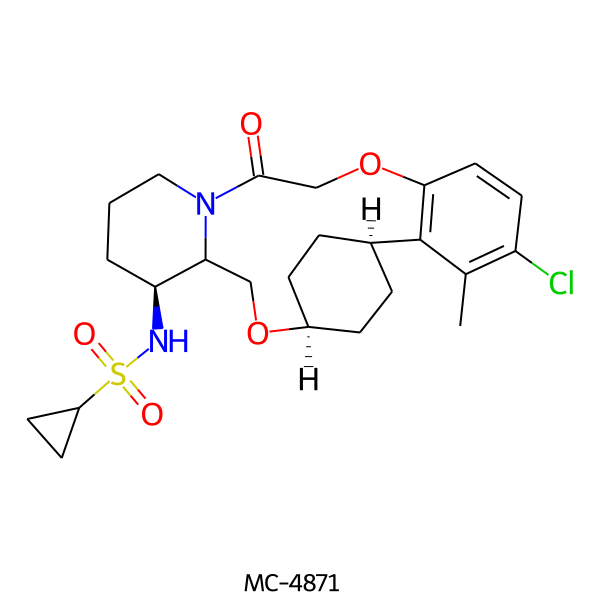 |
497.06 | 5 | 1 | 3.53 | 93.32 | 6.64 | |
| MC-4872 | 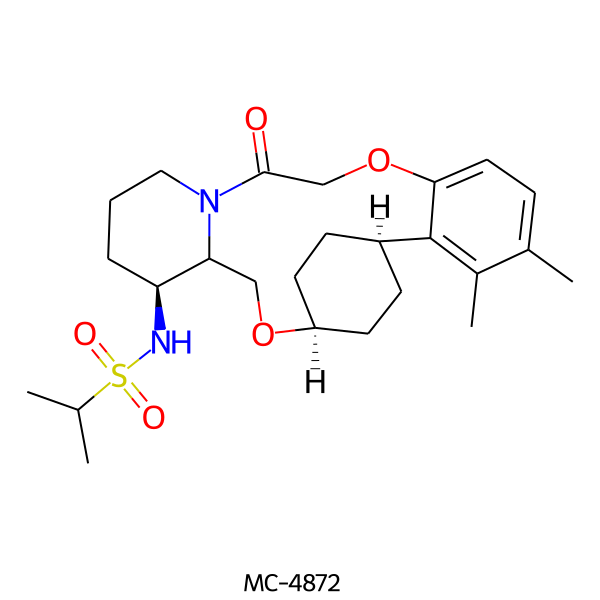 |
478.66 | 5 | 1 | 3.43 | 93.32 | 7.36 | |
| MC-4873 | 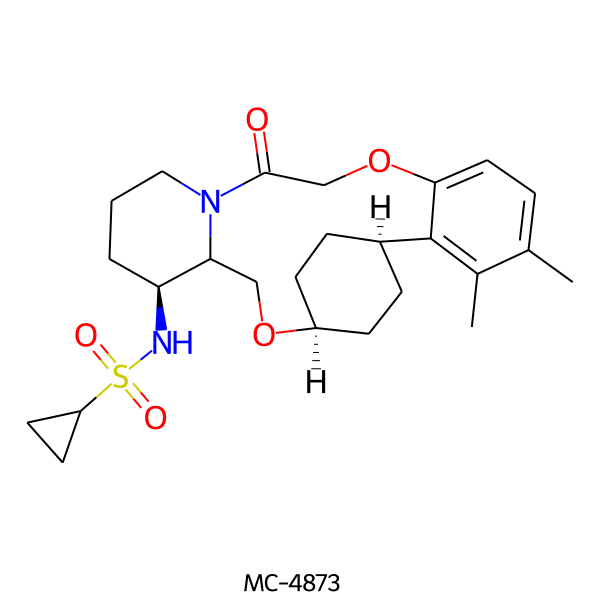 |
476.64 | 5 | 1 | 3.18 | 93.32 | 6.44 | |
| MC-4874 | 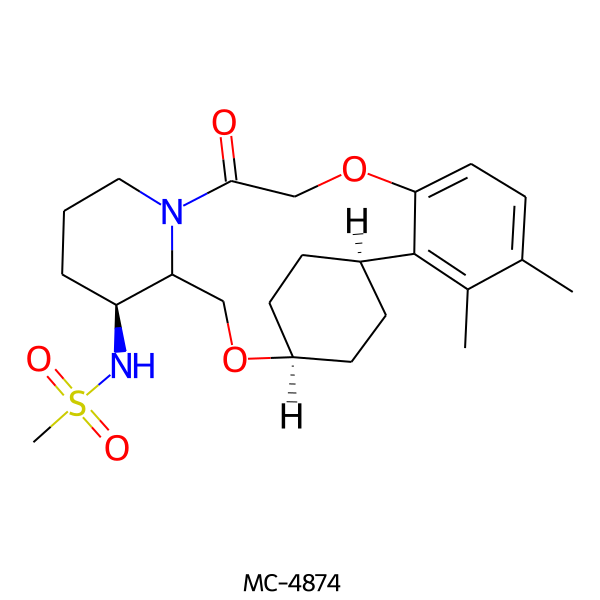 |
450.60 | 5 | 1 | 2.65 | 93.32 | 6.60 | |
| MC-4875 | 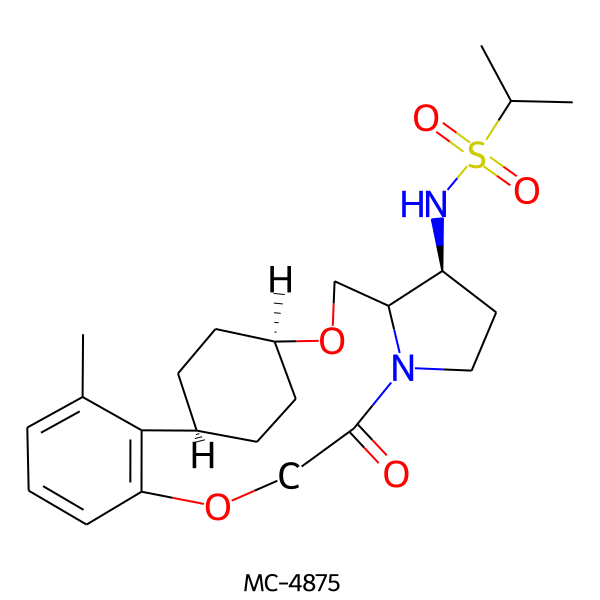 |
450.60 | 5 | 1 | 2.73 | 93.32 | 6.60 | |
| MC-4876 | 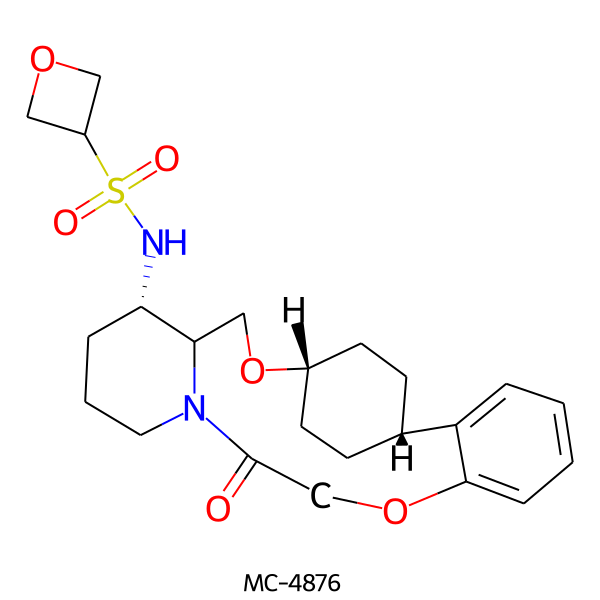 |
464.58 | 6 | 1 | 1.80 | 102.55 | 6.42 | |
| MC-4877 | 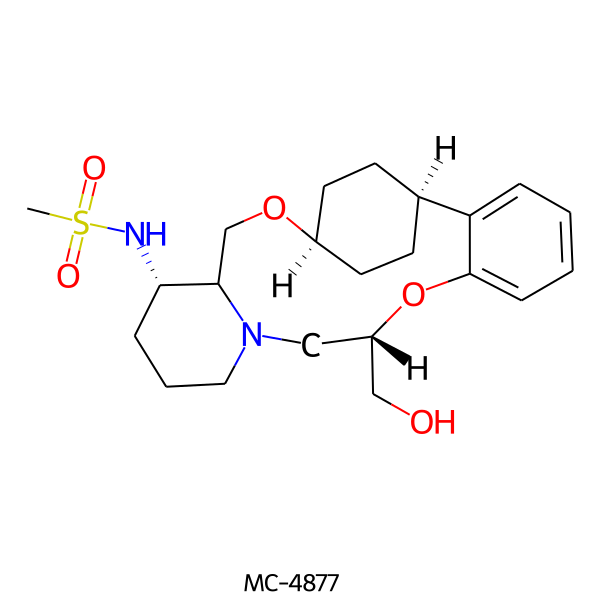 |
438.59 | 6 | 2 | 1.86 | 96.48 | 6.99 | |
| MC-4878 | 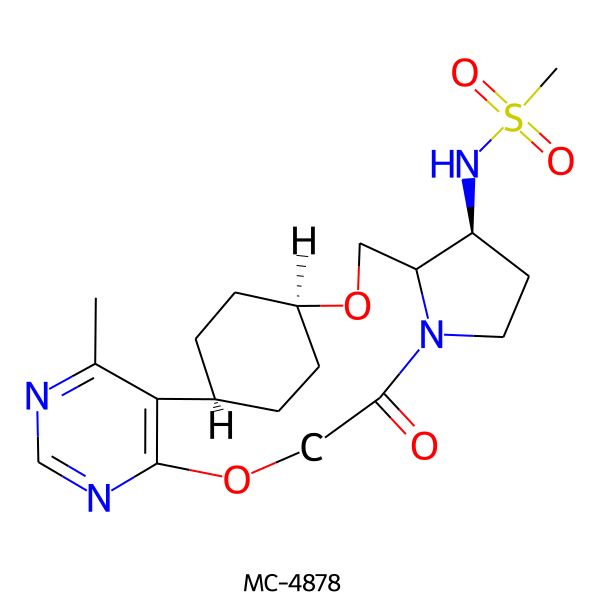 |
424.52 | 7 | 1 | 0.74 | 119.10 | 5.76 | |
| MC-4879 | 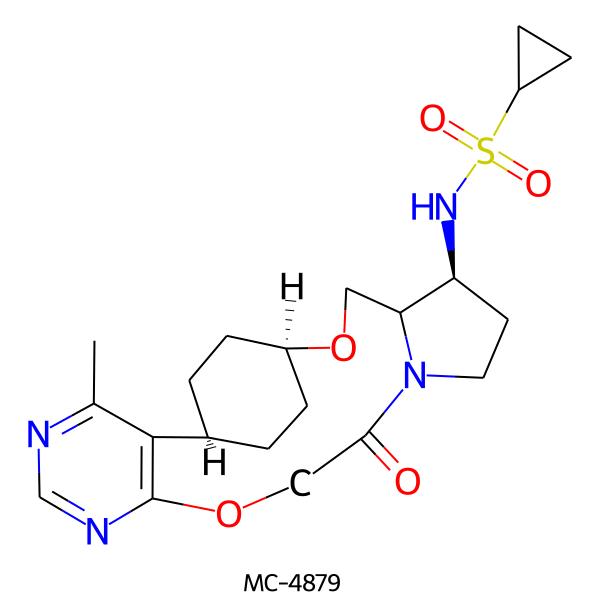 |
450.56 | 7 | 1 | 1.27 | 119.10 | 5.64 | |
| MC-4880 | 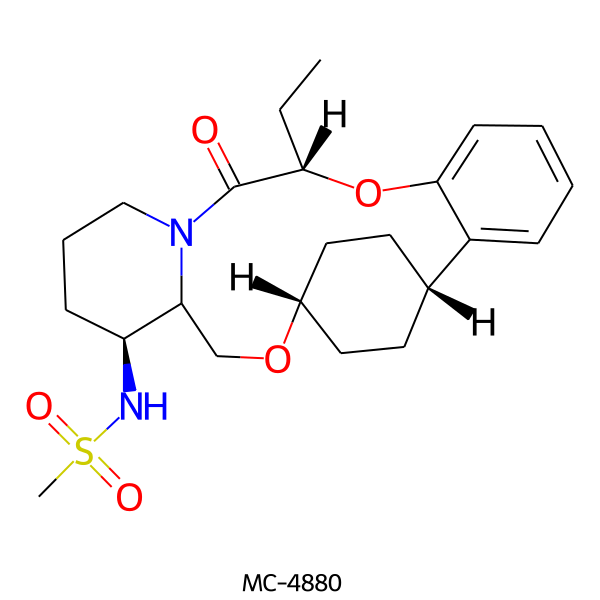 |
450.60 | 5 | 1 | 2.81 | 93.32 | 6.89 | |
| MC-4881 | 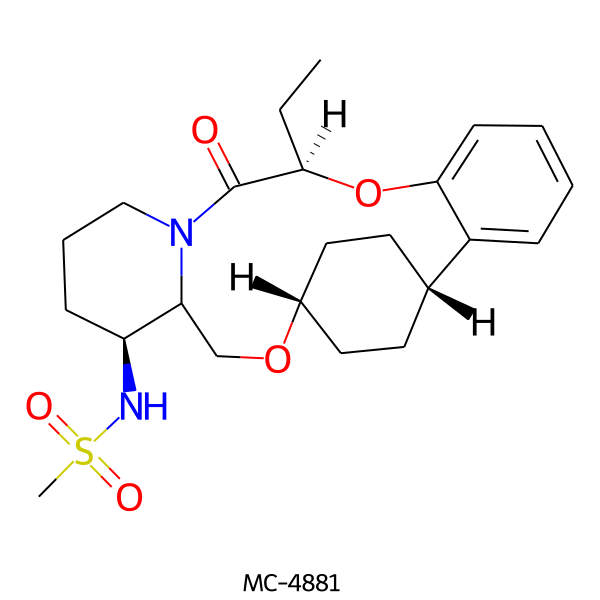 |
450.60 | 5 | 1 | 2.81 | 93.32 | 6.89 | |
| MC-4882 | 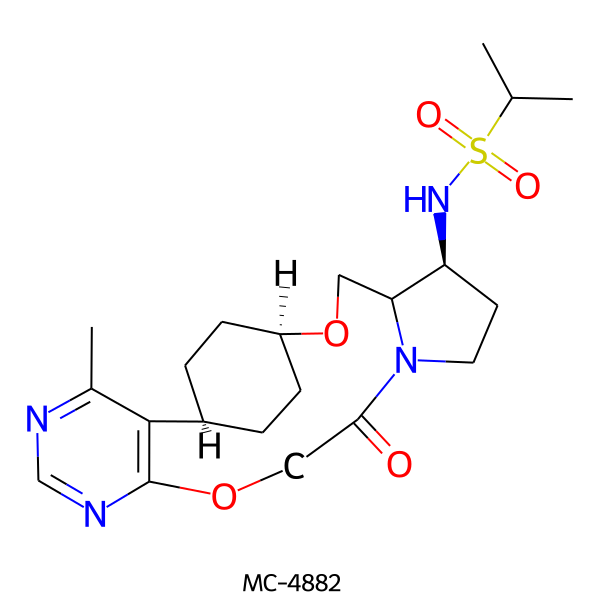 |
452.58 | 7 | 1 | 1.52 | 119.10 | 6.50 | |
| MC-4883 | 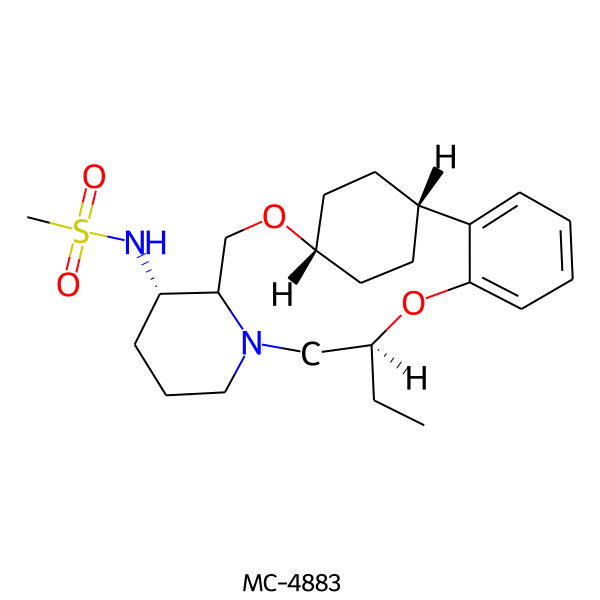 |
436.62 | 5 | 1 | 3.28 | 76.25 | 7.02 | |
| MC-4884 | 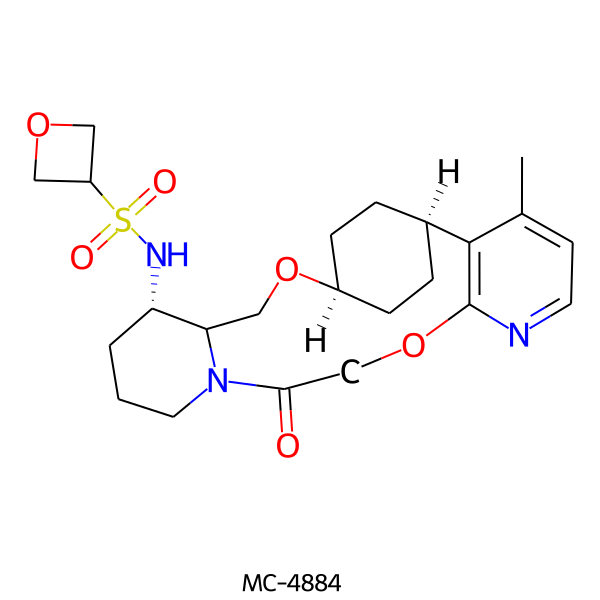 |
479.60 | 7 | 1 | 1.50 | 115.44 | 6.61 | |
| MC-4885 | 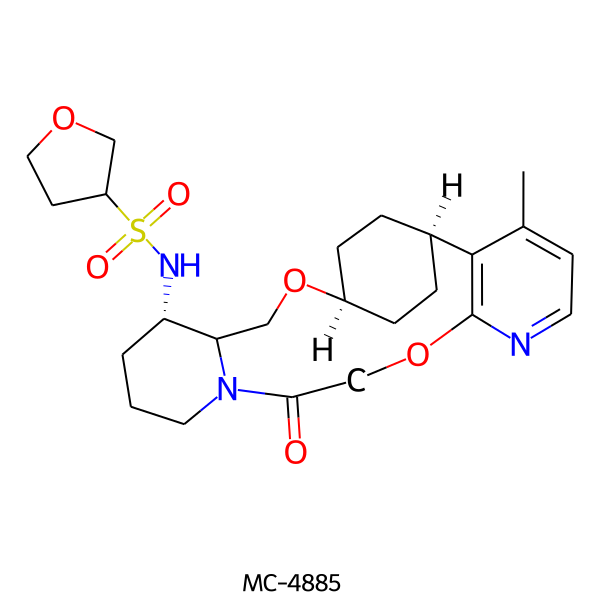 |
493.63 | 7 | 1 | 1.89 | 115.44 | 7.11 | |
| MC-4886 | 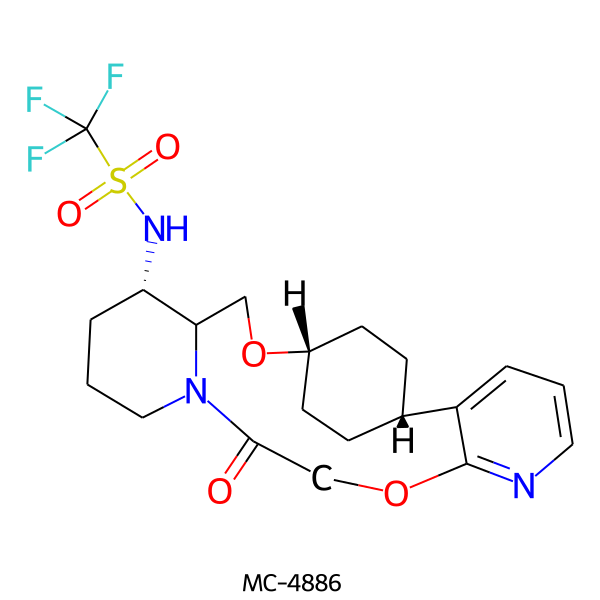 |
477.50 | 6 | 1 | 2.32 | 106.21 | 6.64 | |
| MC-4887 | 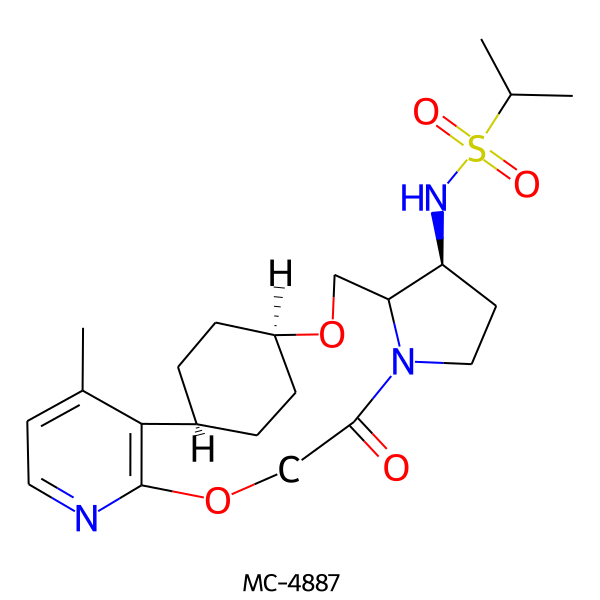 |
451.59 | 6 | 1 | 2.12 | 106.21 | 6.55 | |
| MC-4888 | 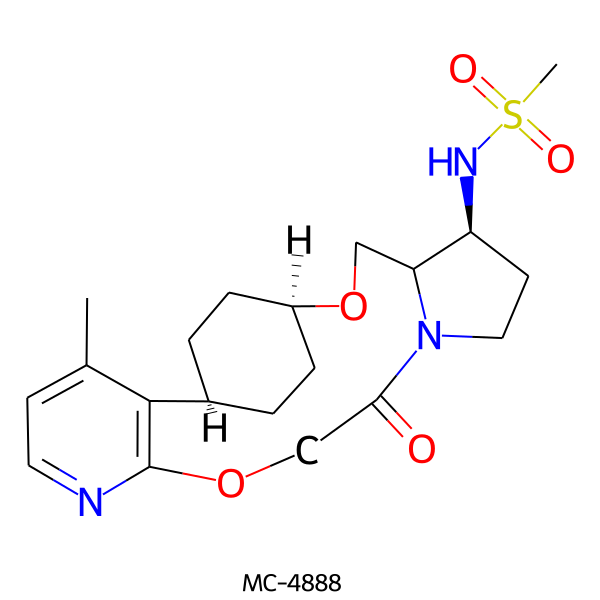 |
423.54 | 6 | 1 | 1.34 | 106.21 | 5.81 | |
| MC-4889 | 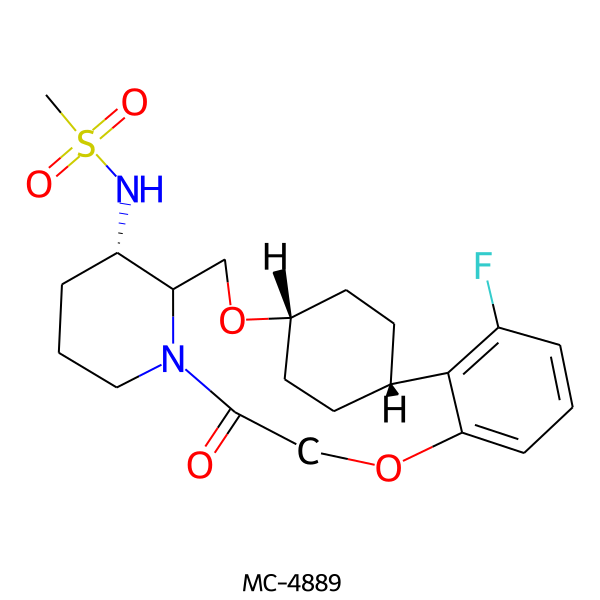 |
440.54 | 5 | 1 | 2.17 | 93.32 | 6.31 | |
| MC-4890 | 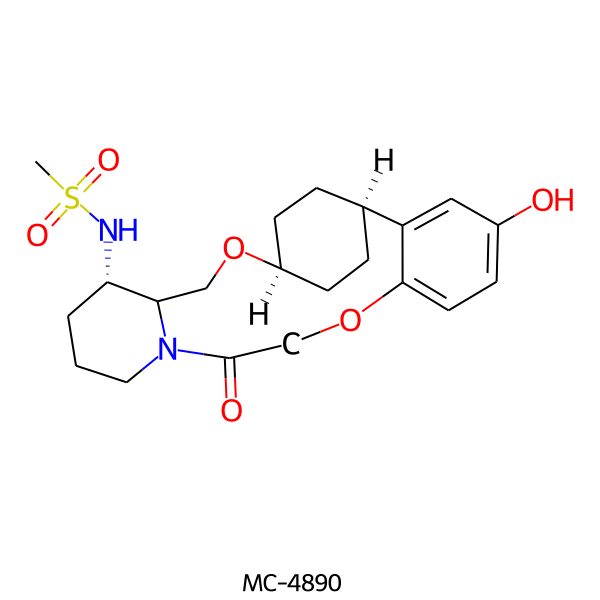 |
438.55 | 6 | 2 | 1.74 | 113.55 | 6.22 | |
| MC-4891 | 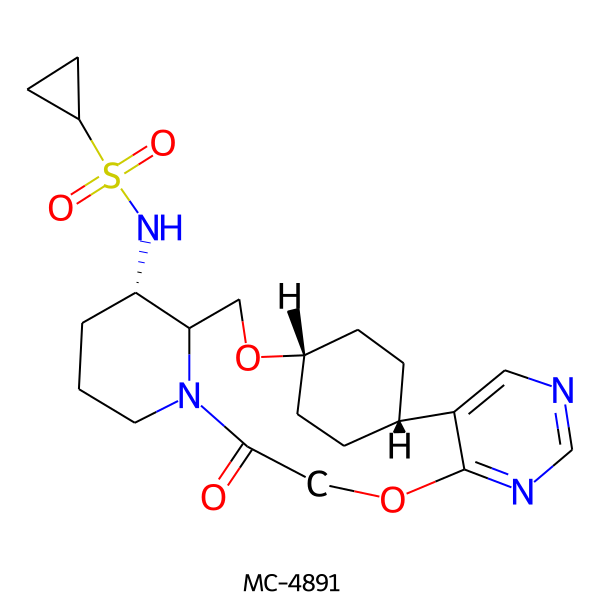 |
450.56 | 7 | 1 | 1.35 | 119.10 | 5.87 | |
| MC-4892 | 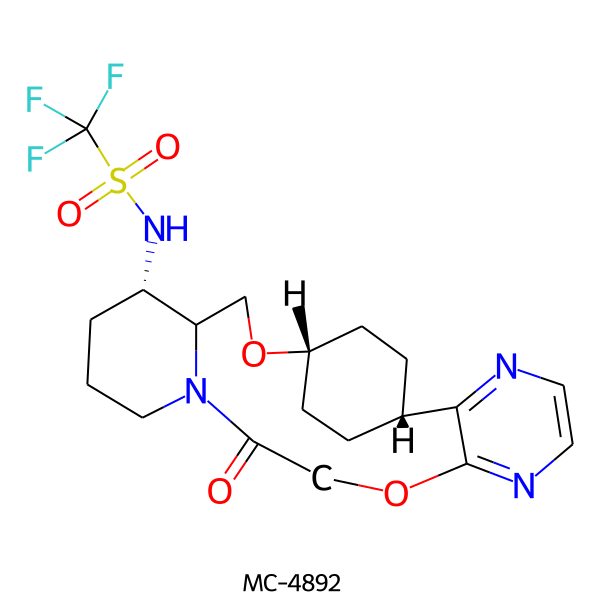 |
478.49 | 7 | 1 | 1.71 | 119.10 | 6.59 | |
| MC-4893 | 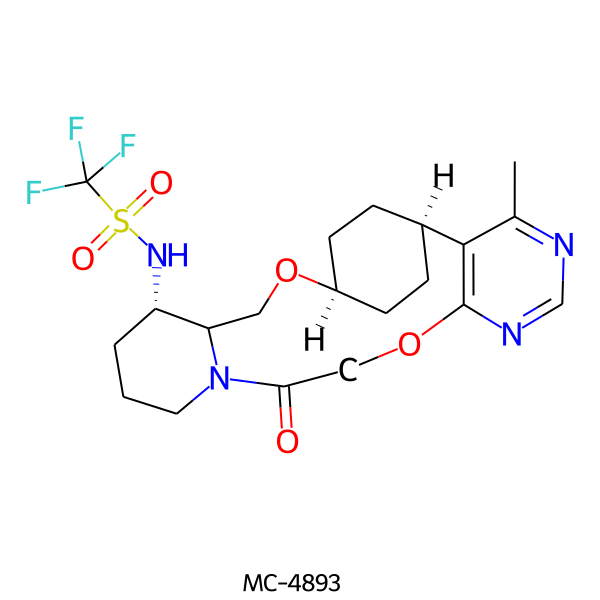 |
492.52 | 7 | 1 | 2.02 | 119.10 | 6.83 | |
| MC-4894 | 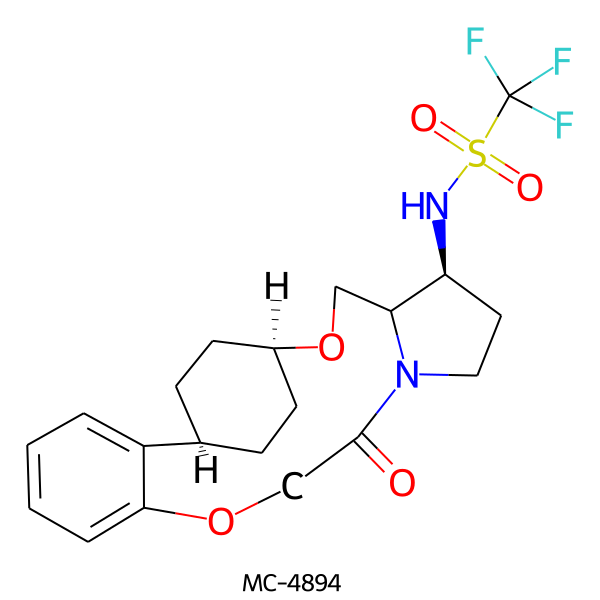 |
462.49 | 5 | 1 | 2.53 | 93.32 | 6.19 | |
| MC-4895 | 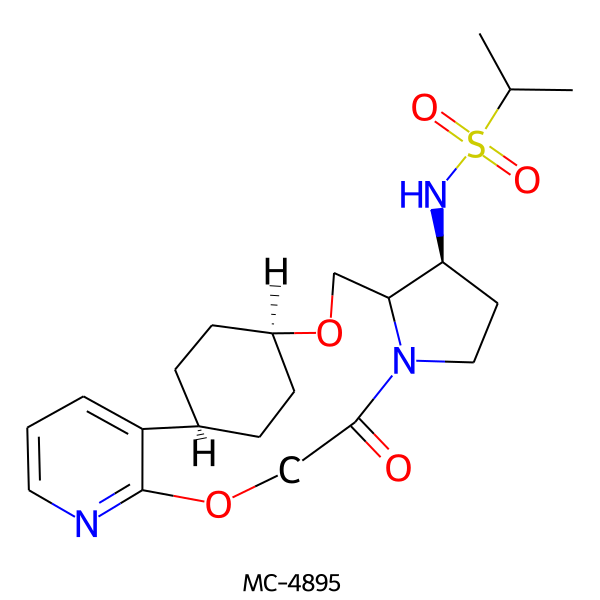 |
437.56 | 6 | 1 | 1.81 | 106.21 | 6.31 | |
| MC-4896 | 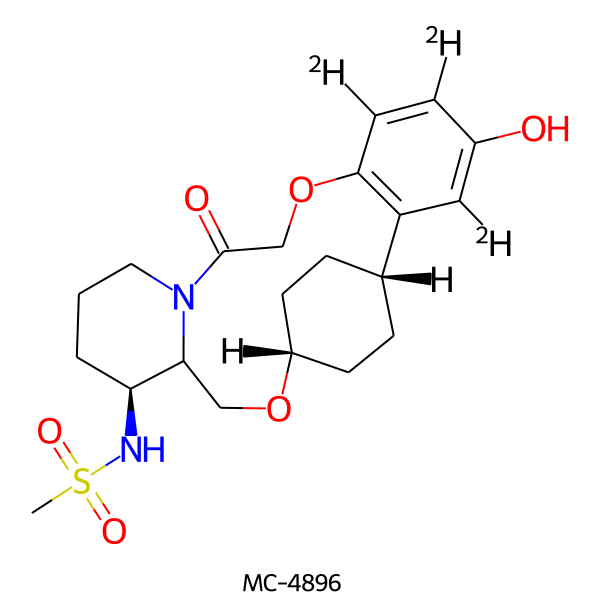 |
441.56 | 6 | 2 | 1.74 | 113.55 | 5.18 | |
| MC-4897 | 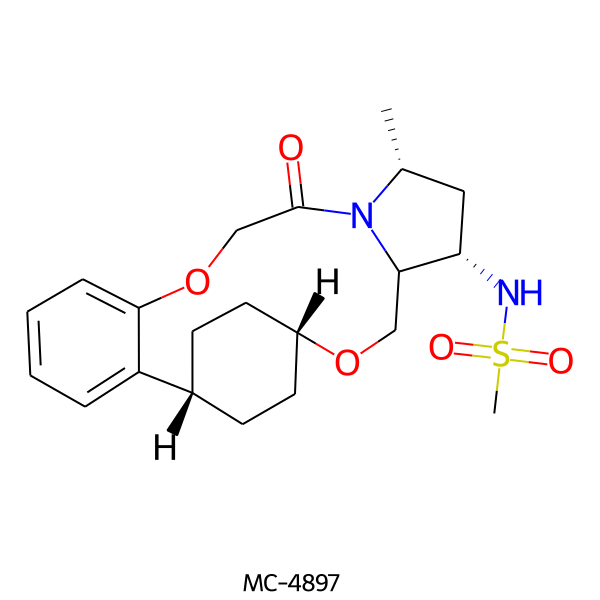 |
422.55 | 5 | 1 | 2.03 | 93.32 | 5.86 | |
| MC-4898 | 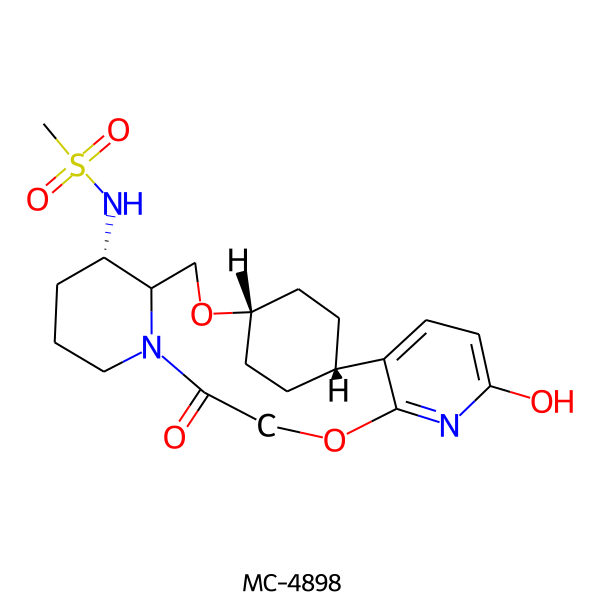 |
439.53 | 7 | 2 | 1.13 | 126.44 | 6.17 | |
| MC-4899 | 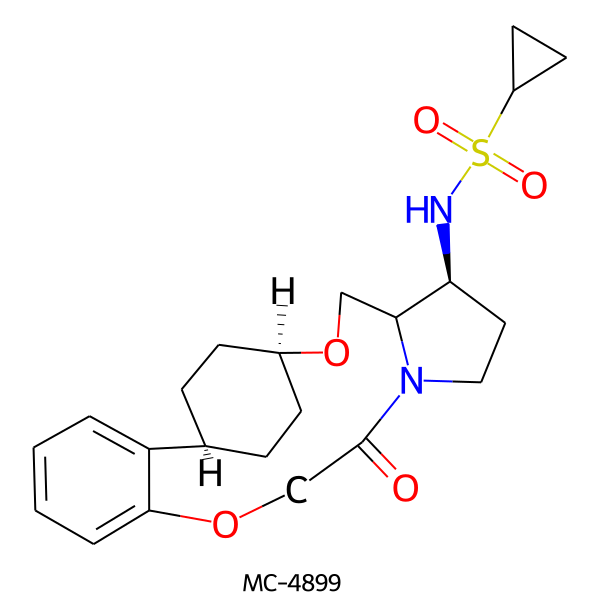 |
434.56 | 5 | 1 | 2.17 | 93.32 | 5.49 |
| Select | Name | Assay | Endpoint | Value | Unit | Standardized Value | Source |
|---|---|---|---|---|---|---|---|
| MC-0082 | MDCK | Papp AB | 6.8 | 10-6 cm/s | -5.17 | (Pennington et al., 2021) | |
| MC-0082 | MDCK | ER | 0.75 | 0.75 | (Pennington et al., 2021) | ||
| MC-0082 | MDCK | Papp AB | 9.1 | 10-6 cm/s | -5.04 | (Pennington et al., 2021) |
Back to Browse