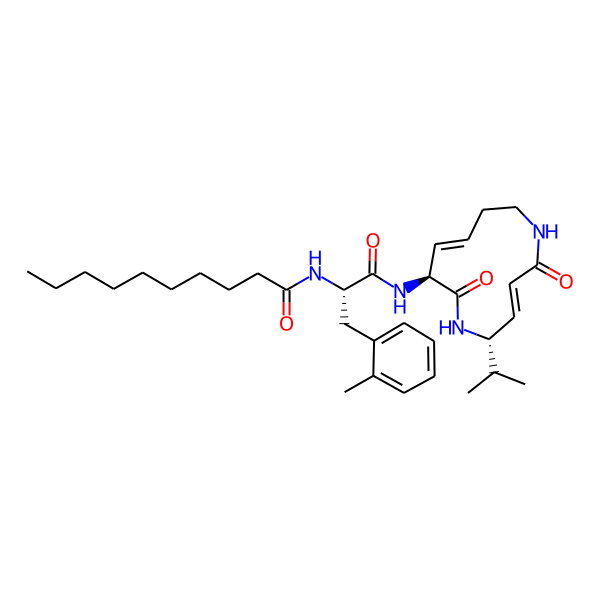
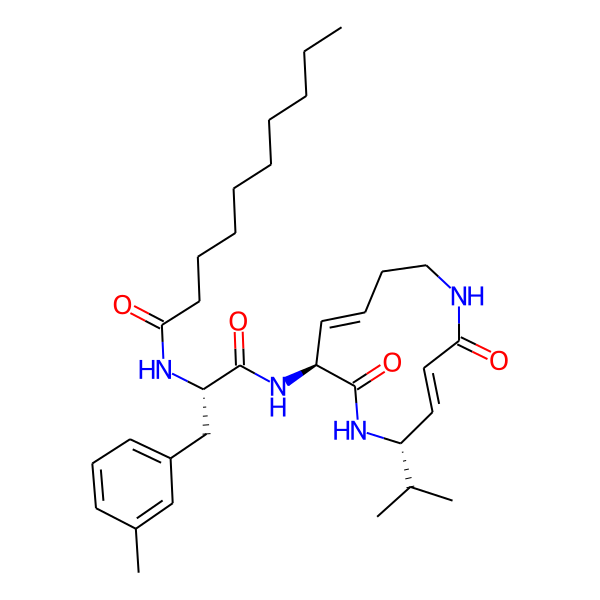
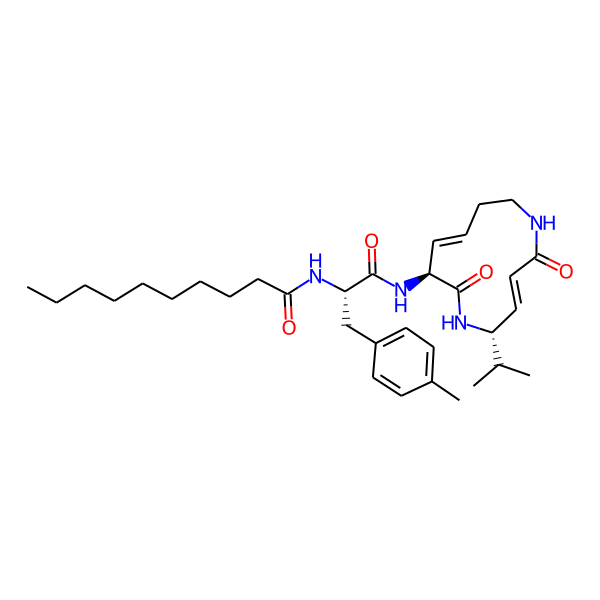
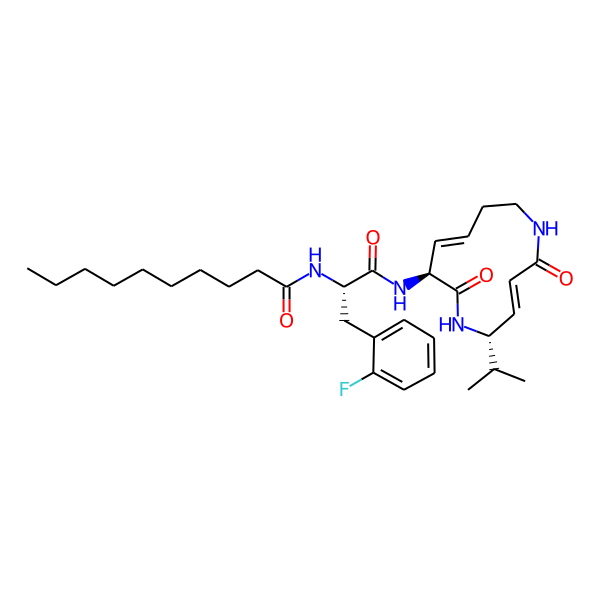
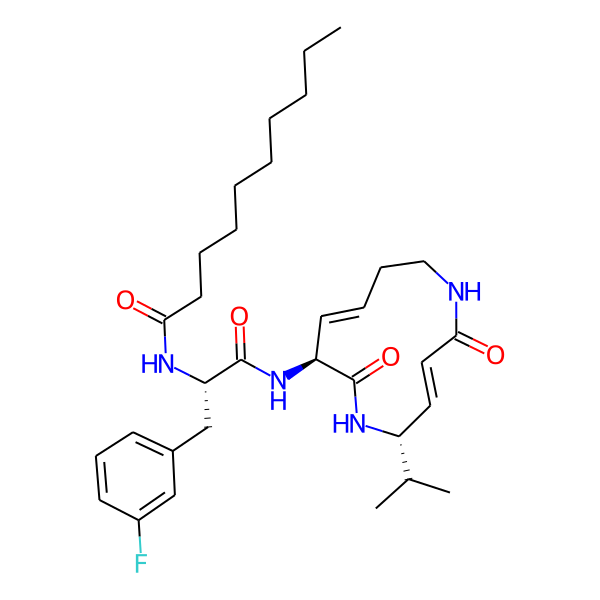
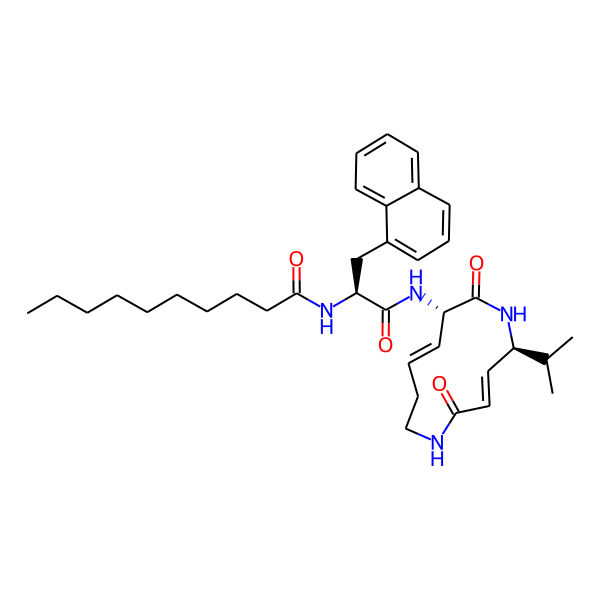
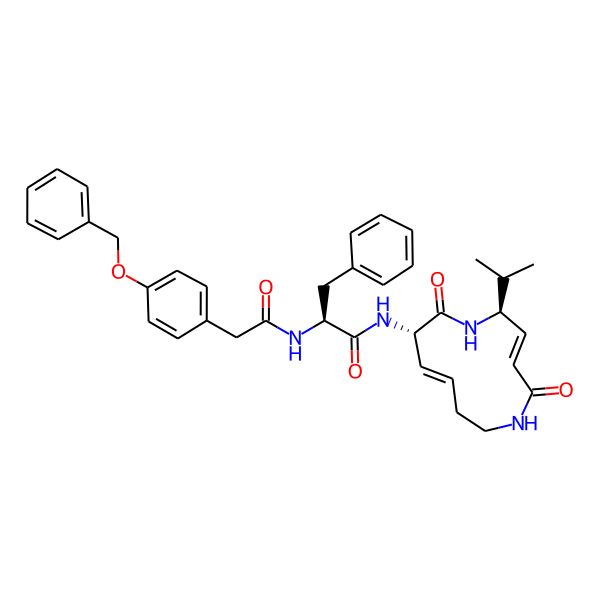
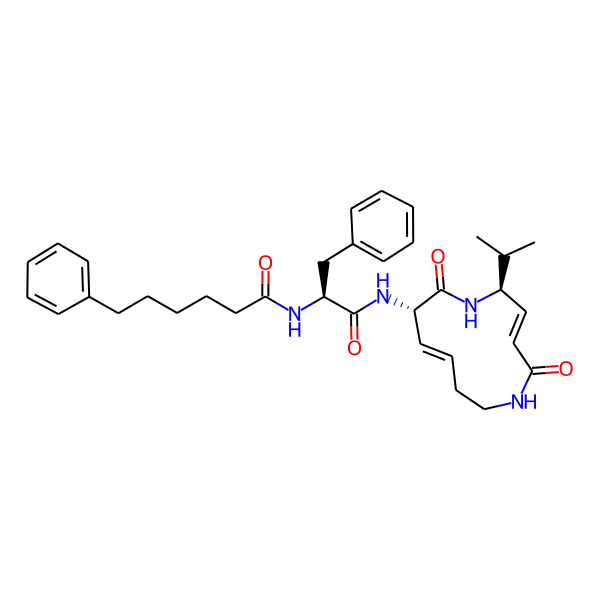
MC-3800
| Name | |||
|---|---|---|---|
| Unique ID | MC-3800 | ||
| Original ID | HU9-281 (Miyachi et al., 2021) | ||
| Common Name | |||
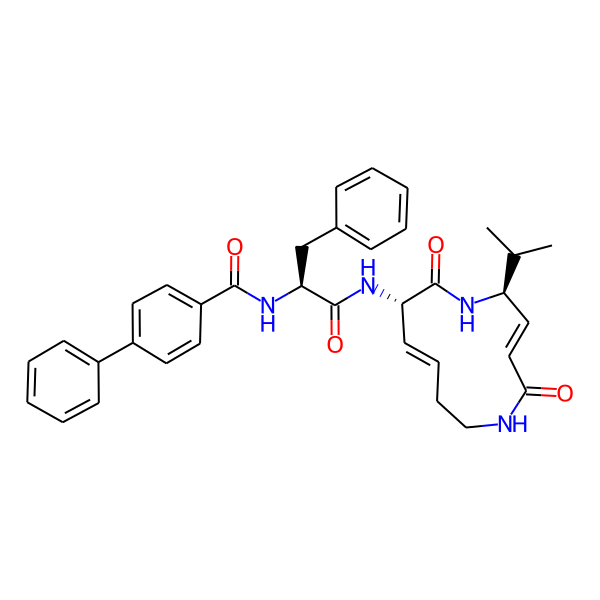
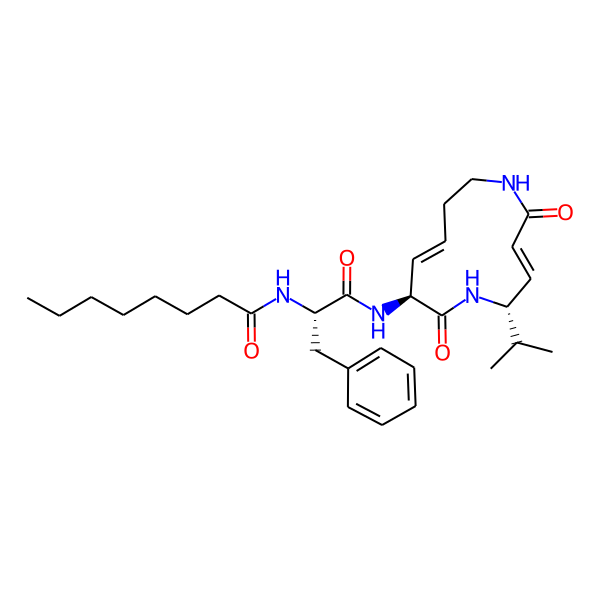
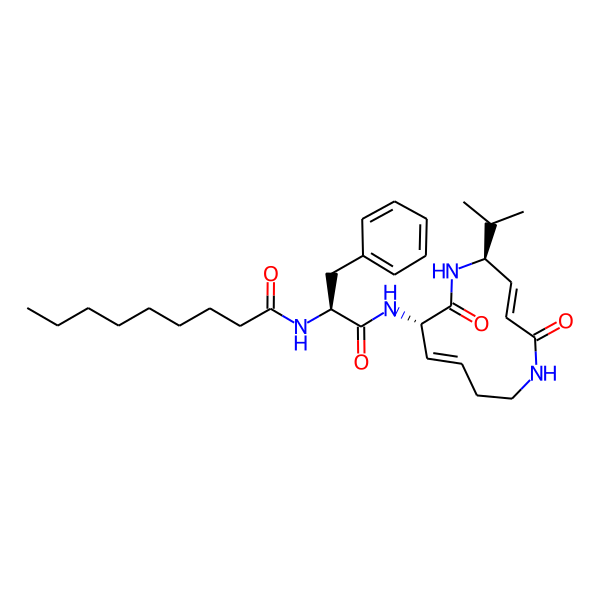
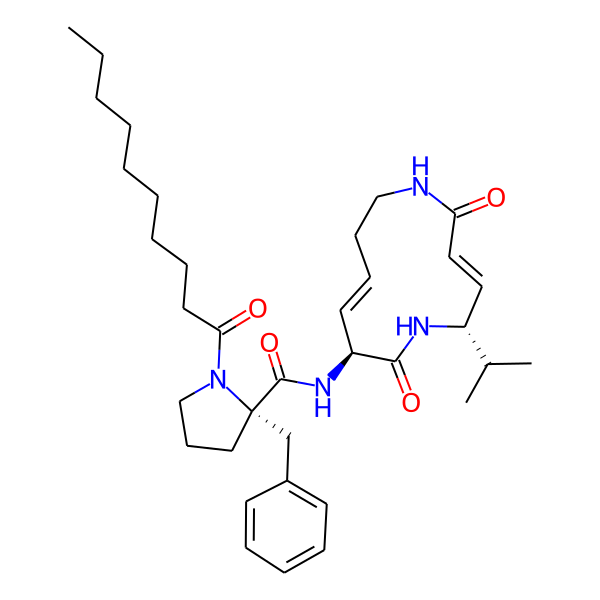
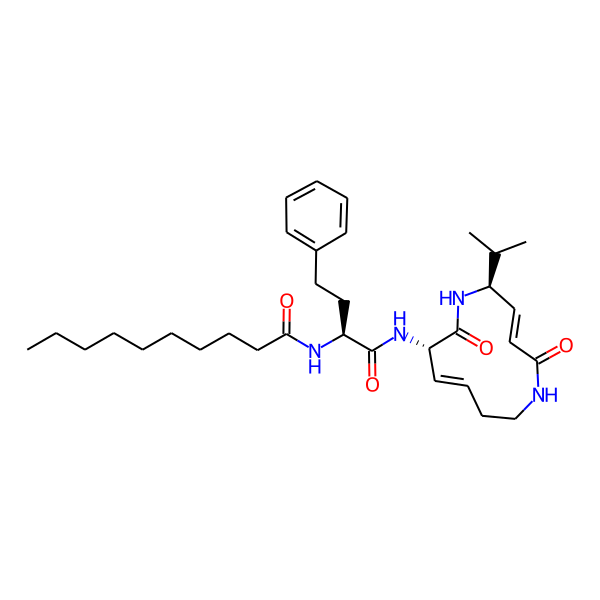
| Structure Representations | |||
| InchiKey | SDVZKHGXBJTNSJ-SLZDWNPMSA-N | ||
| Isomeric SMILES | CC(C)[C@H]1/C=C/C(=O)NCC/C=C/[C@H](NC(=O)[C@H](Cc2ccccc2)NC(=O)c2ccc(-c3ccccc3)cc2)C(=O)N1 | ||
| SMILES (Ring) | C1=CCCNCC=CCNCC1 | ||
| Permeability | |||
| Assay | PAMPA | ||
| Endpoint | Papp | ||
| Value | 0.063 | ||
| Unit | 10-6 cm/s | ||
| Standardized Value | -7.20 | ||
| Molecule Descriptors | |||
| MW (Da) | 578.71 | NRotB | 8 |
| HBA | 4 | Kier Index (Φ) | 11.02 |
| HBD | 4 | AR | 0.50 |
| cLogP | 3.95 | Fsp3 | 0.26 |
| TPSA (Å2) | 116.40 | MRS | 12 |
| Unique ID: ID for each unique molecule in this dataset; Original ID: The name or ID in original source; Standaridized value: Logarithmic values for permeability values (unit 10-6 cm/s), original data with sign '<' or '>' were removed during data standardization | |||
| Abbreviations: MW (Da): Molecular Weight; HBA: Hydrogen bond acceptor; HBD: Hydrogen bond donor; cLogP: Calculated lipophilicity; TPSA: Topological polar surface area; NRotB: Number of rotatable bonds; Φ: Kier flexibility Index: Fsp3: fraction of sp3 carbon atoms; MRS: macrocyclic ring size; AR: Amide ratio; | |||
Back to Browse